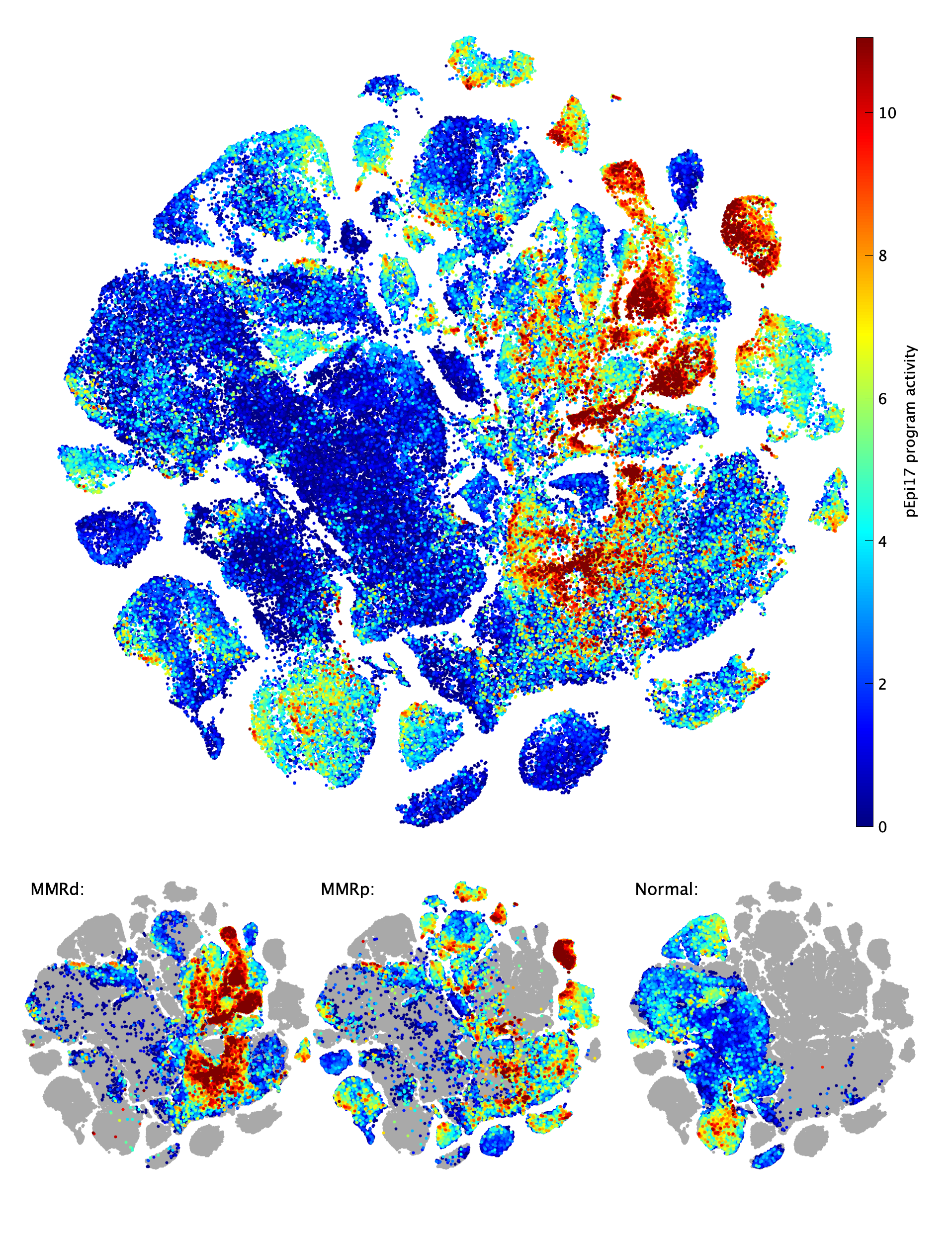
pEpi17 program activity among Epithelial cells
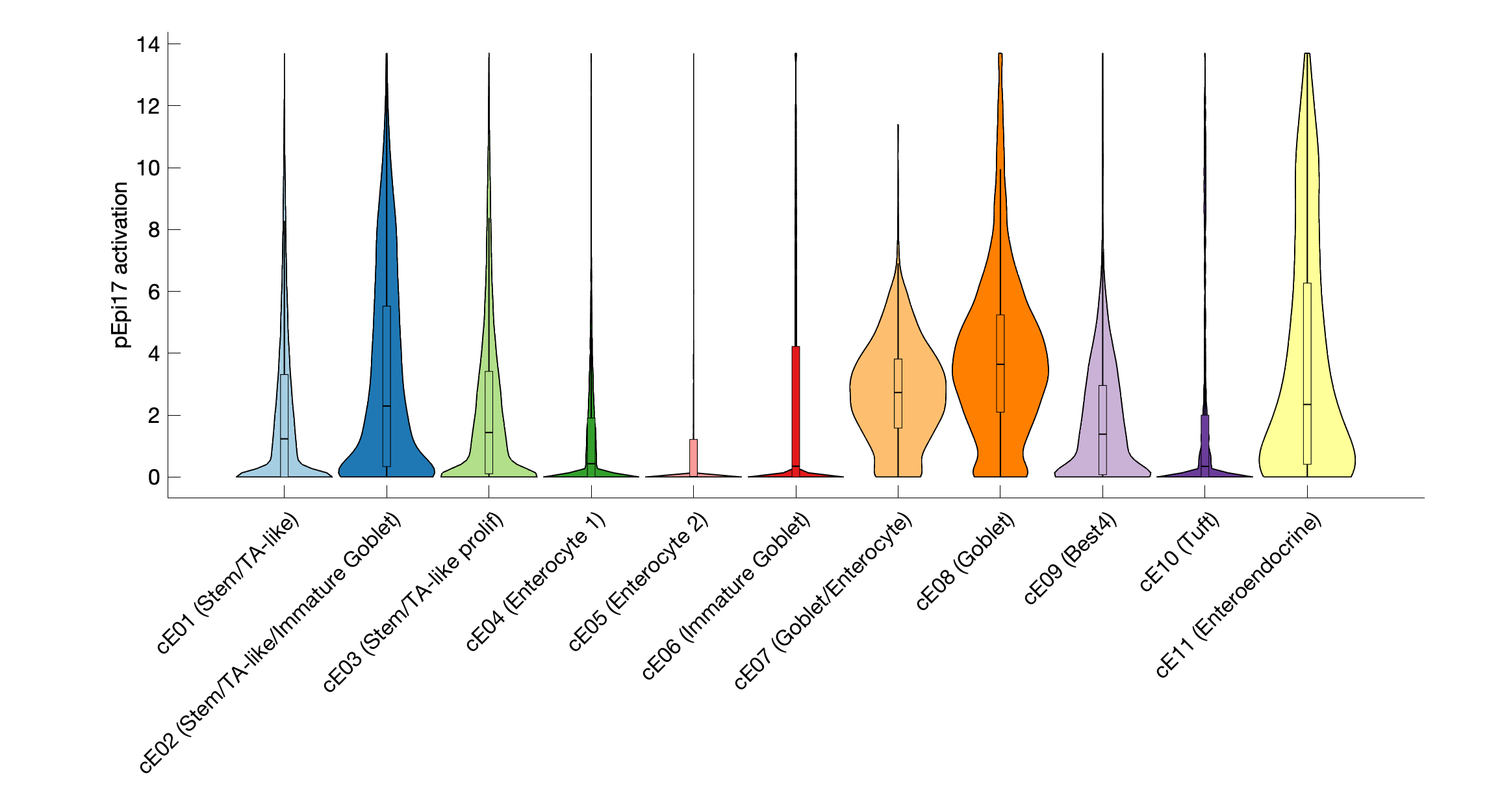
Epithelial cell compositional overview

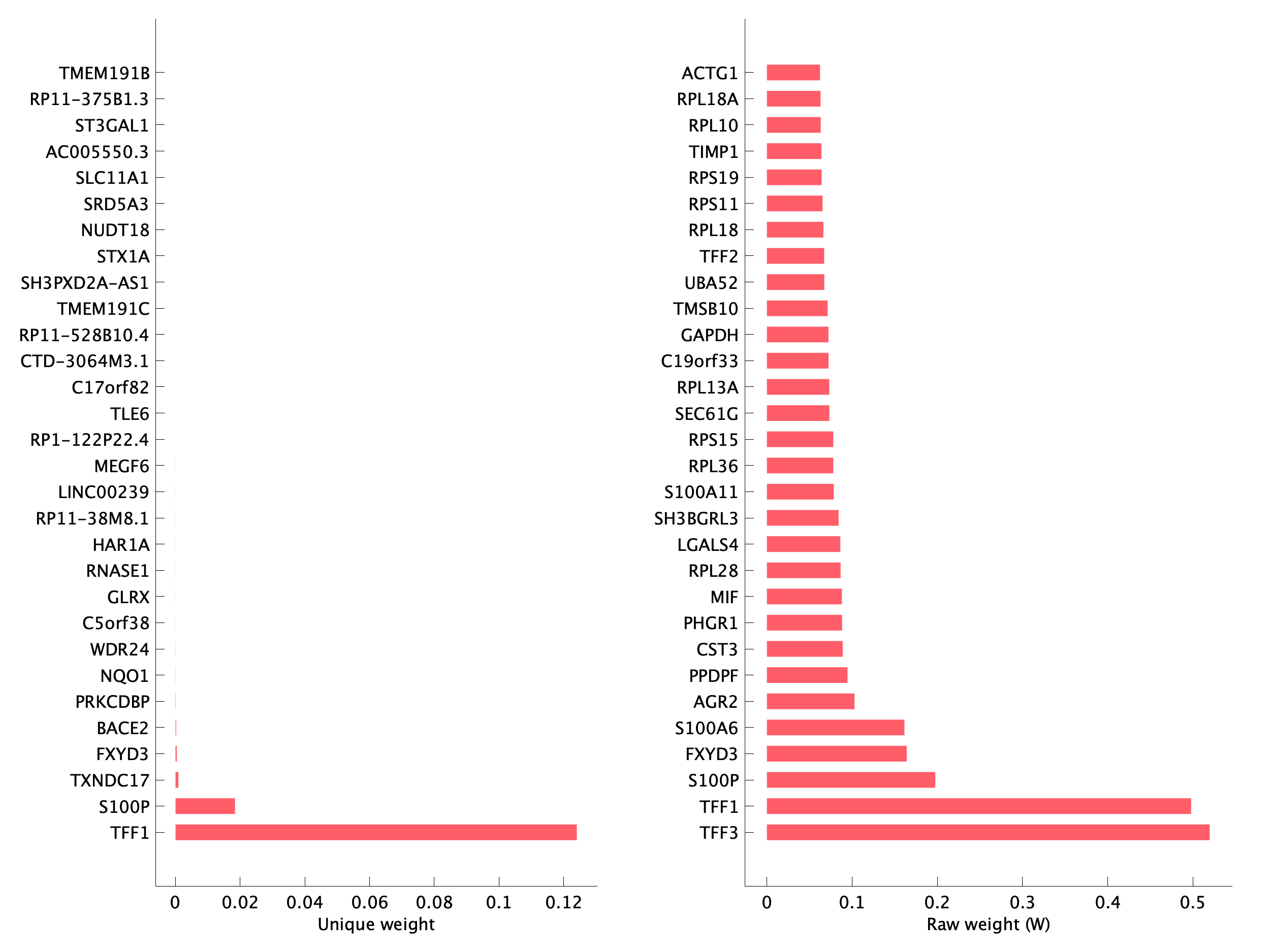
pEpi17 top program genes
pEpi17 gene-set enrichment of top program genes (using g-profiler)
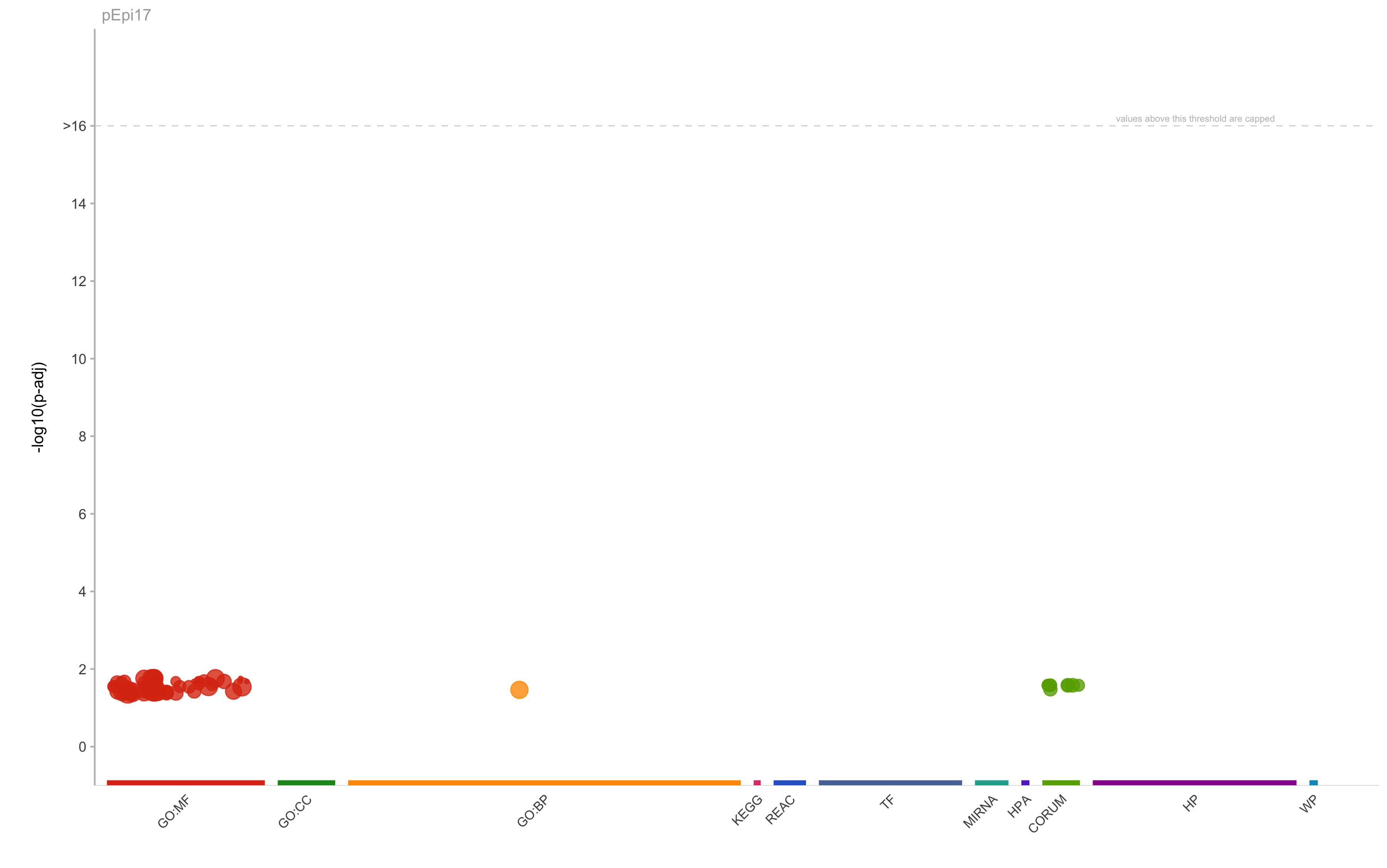
| Source | termID | Name | Padj | Tsize | Qsize | Overlap | Precision | Recall |
|---|---|---|---|---|---|---|---|---|
| GO:BP | GO:0045454 | cell redox homeostasis | 0.0344178244270067 | 58 | 19 | 3 | 0.157894736842105 | 0.0517241379310345 |
| GO:MF | GO:0048306 | calcium-dependent protein binding | 0.0175091025915747 | 82 | 30 | 3 | 0.1 | 0.0365853658536585 |
| GO:MF | GO:0016491 | oxidoreductase activity | 0.0287749065186658 | 771 | 9 | 3 | 0.333333333333333 | 0.00389105058365759 |
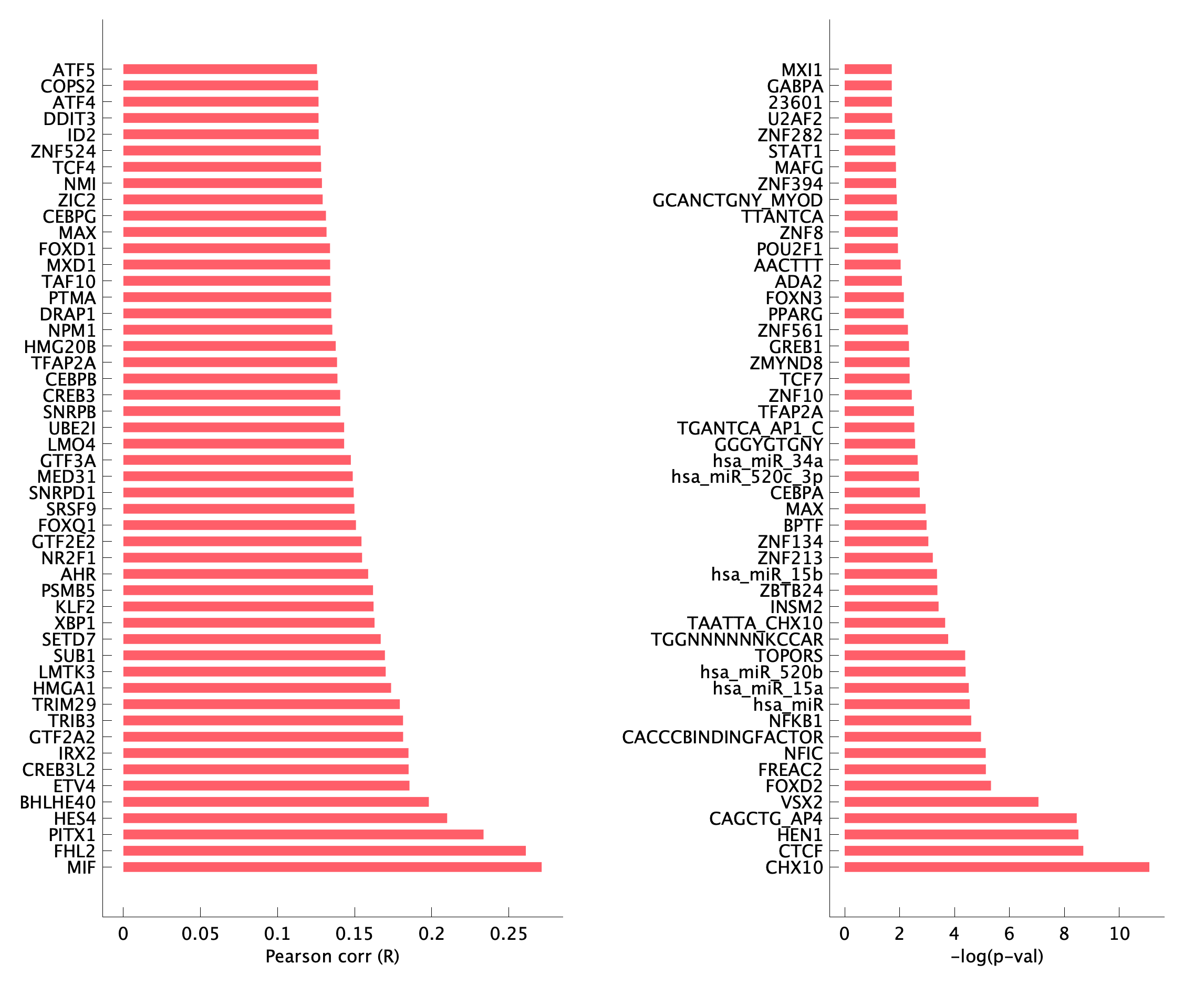
pEpi17 transcription factor target enrichment of top program genes