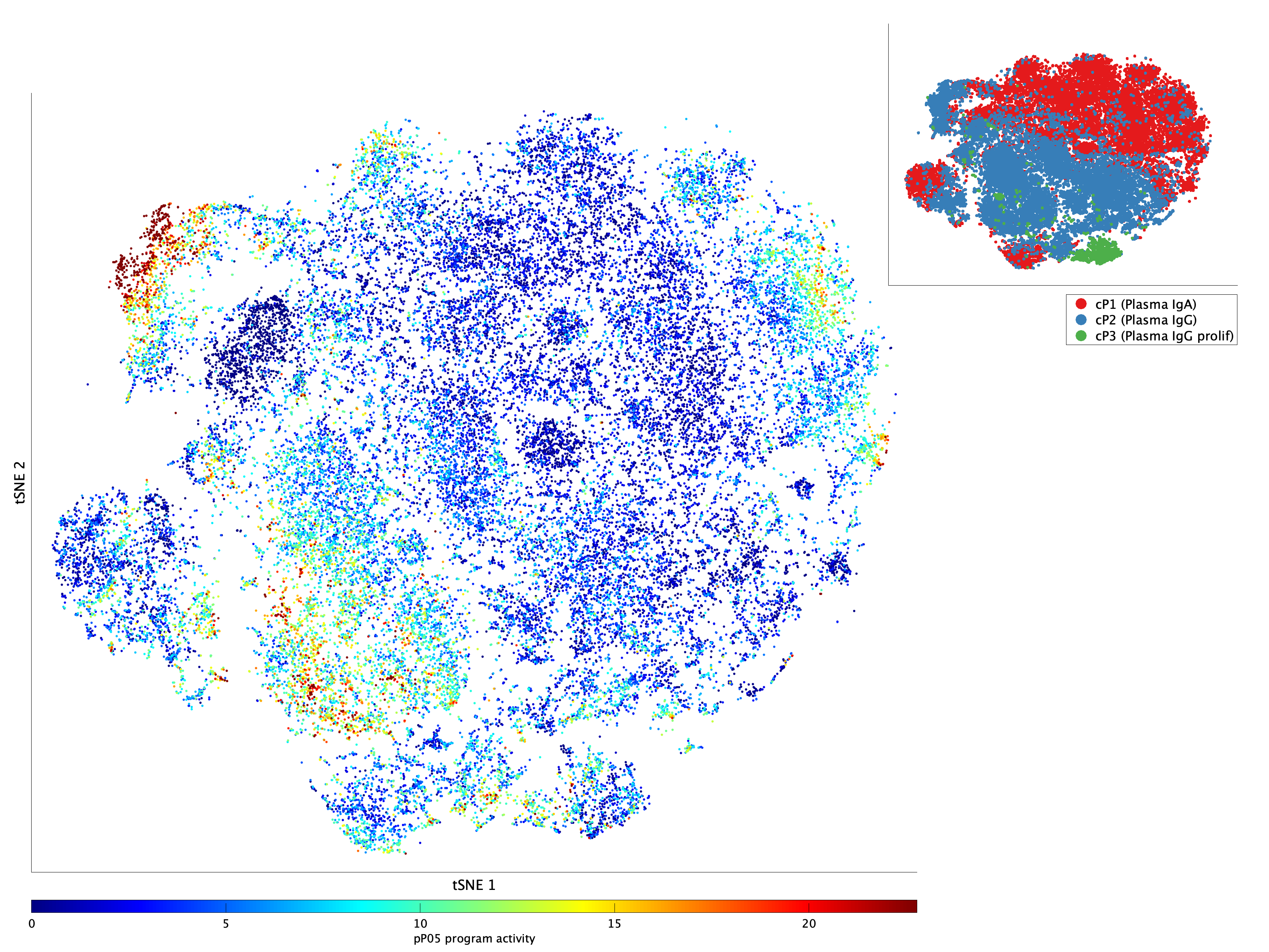
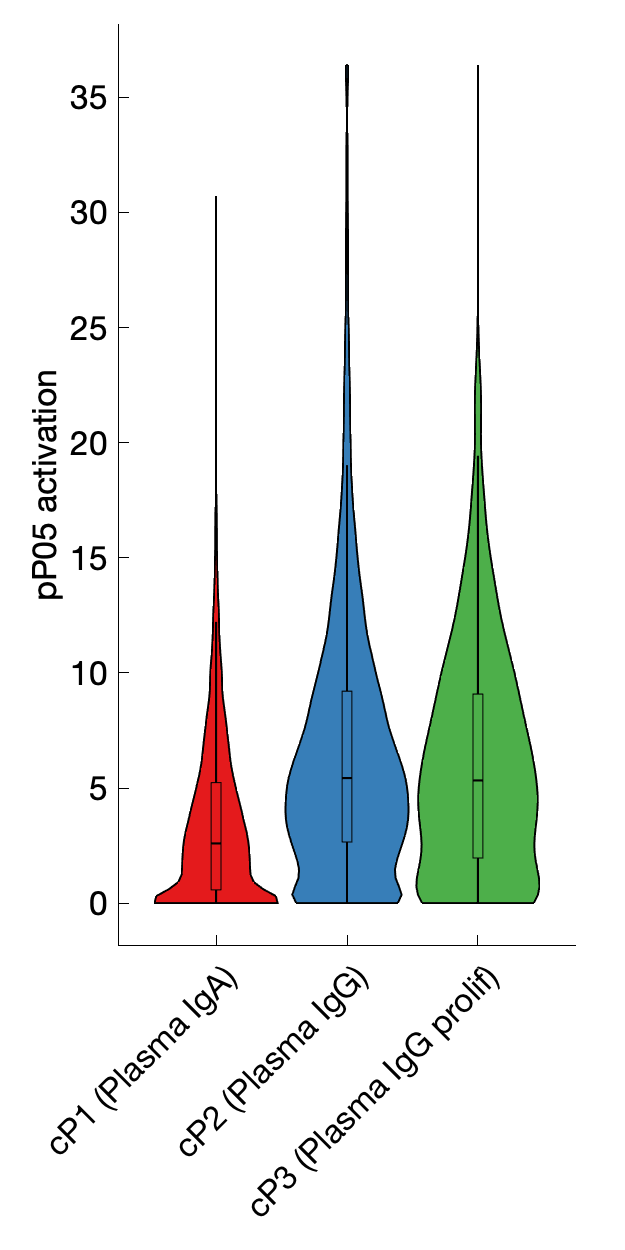
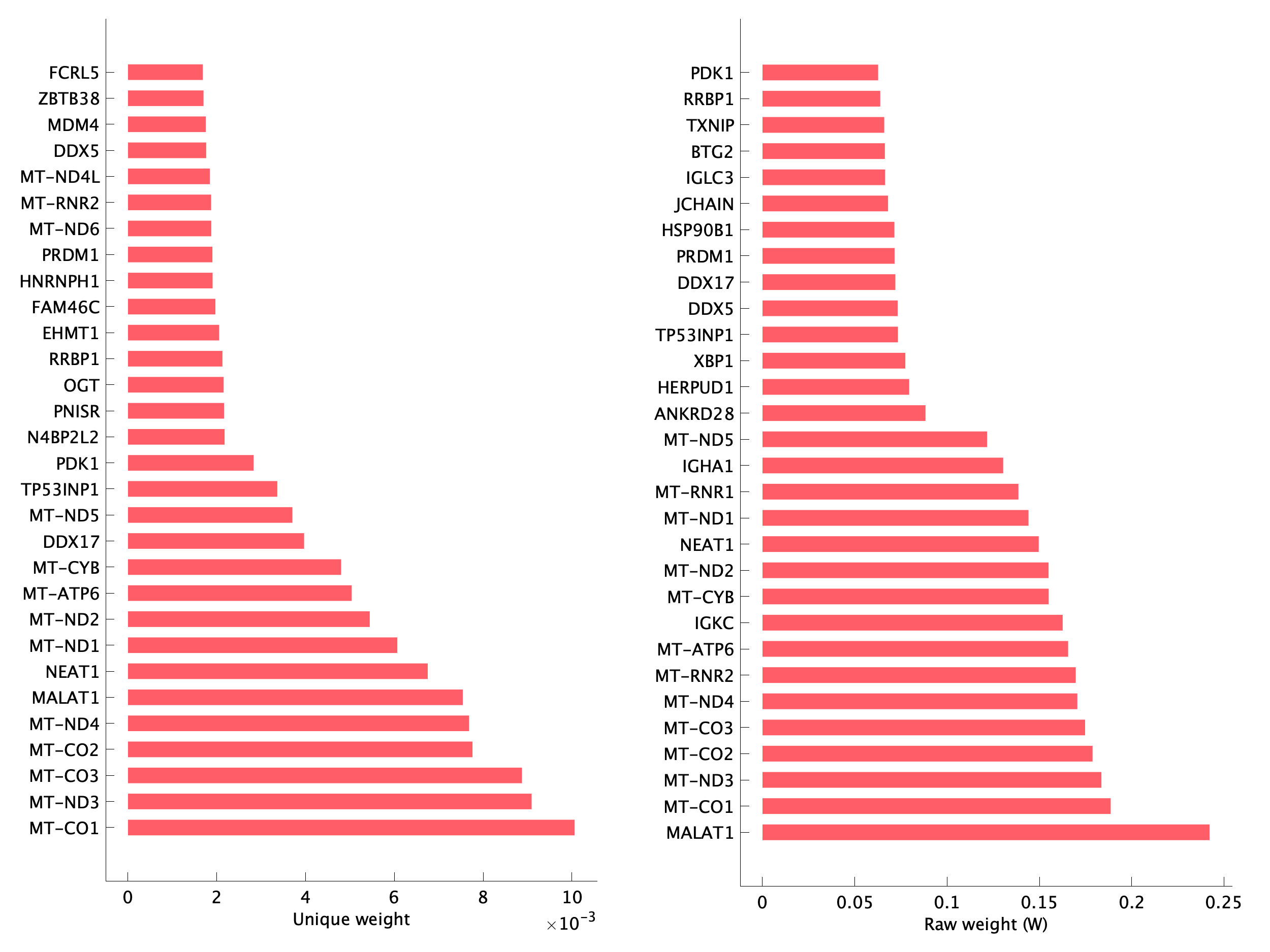
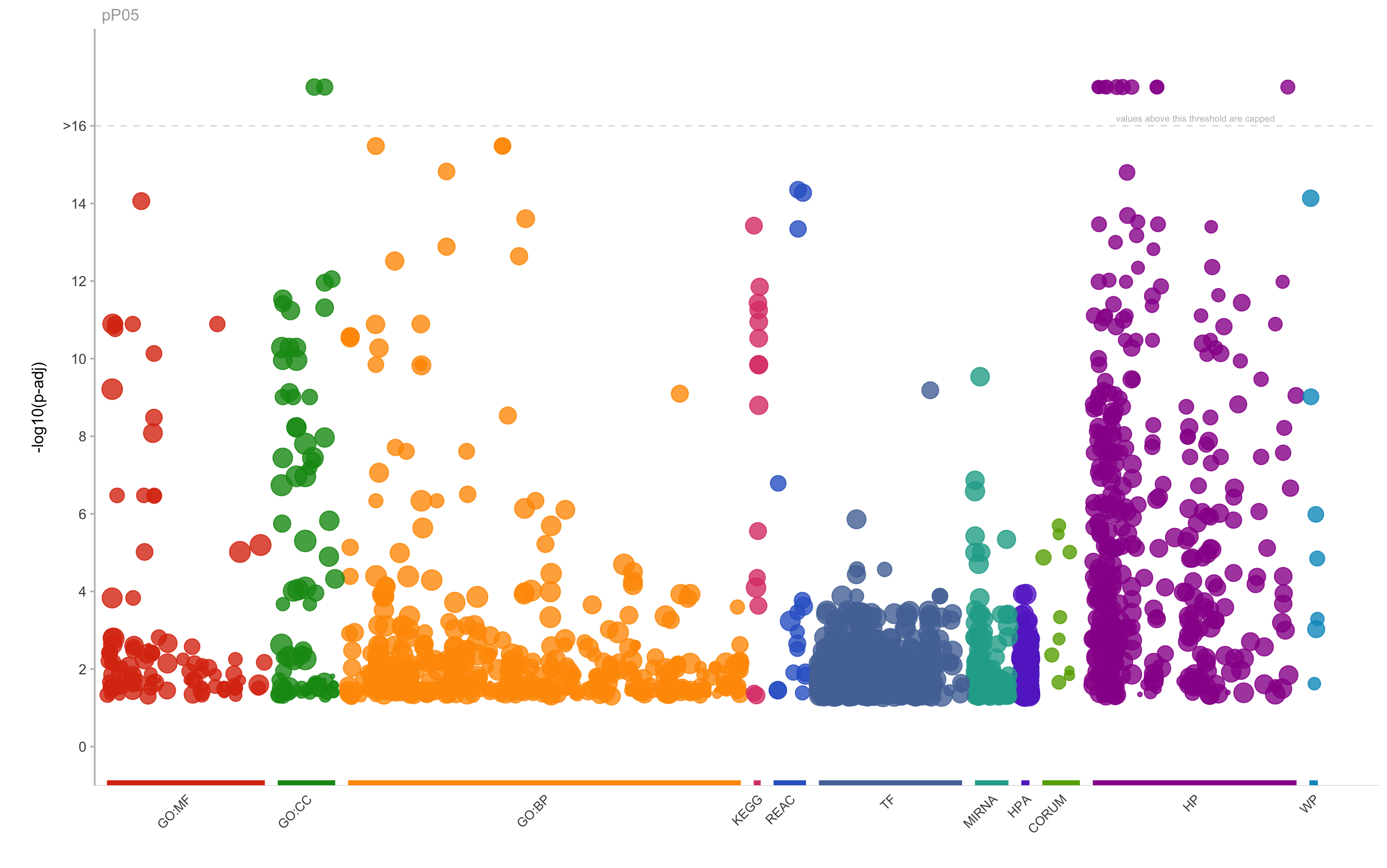
|
CORUM
|
CORUM:2919
|
Respiratory chain complex I (gamma subunit) mitochondrial
|
2.00875428816888e-06
|
13
|
25
|
5
|
0.2
|
0.384615384615385
|
|
CORUM
|
CORUM:2886
|
Respiratory chain complex I (incomplete intermediate ND1, ND2, ND3, CIA30 assembly), mitochondrial
|
3.35764426844288e-06
|
4
|
9
|
3
|
0.333333333333333
|
0.75
|
|
CORUM
|
CORUM:6442
|
Cytochrome c oxidase, mitochondrial
|
9.68981163310404e-06
|
14
|
4
|
3
|
0.75
|
0.214285714285714
|
|
CORUM
|
CORUM:178
|
Respiratory chain complex I (holoenzyme), mitochondrial
|
1.32908445163649e-05
|
44
|
25
|
6
|
0.24
|
0.136363636363636
|
|
CORUM
|
CORUM:3082
|
DGCR8 multiprotein complex
|
0.000459305218853606
|
11
|
55
|
4
|
0.0727272727272727
|
0.363636363636364
|
|
CORUM
|
CORUM:1332
|
Large Drosha complex
|
0.00435121872082373
|
20
|
55
|
4
|
0.0727272727272727
|
0.2
|
|
GO:BP
|
GO:0042773
|
ATP synthesis coupled electron transport
|
3.30616487481925e-16
|
100
|
25
|
11
|
0.44
|
0.11
|
|
GO:BP
|
GO:0042775
|
mitochondrial ATP synthesis coupled electron transport
|
3.30616487481925e-16
|
99
|
25
|
11
|
0.44
|
0.111111111111111
|
|
GO:BP
|
GO:0006119
|
oxidative phosphorylation
|
3.30616487481925e-16
|
151
|
13
|
10
|
0.769230769230769
|
0.0662251655629139
|
|
GO:BP
|
GO:0022904
|
respiratory electron transport chain
|
1.50075497773741e-15
|
117
|
25
|
11
|
0.44
|
0.094017094017094
|
|
GO:BP
|
GO:0046034
|
ATP metabolic process
|
2.47526907817713e-14
|
314
|
25
|
13
|
0.52
|
0.0414012738853503
|
|
GO:BP
|
GO:0022900
|
electron transport chain
|
1.29969506497386e-13
|
180
|
25
|
11
|
0.44
|
0.0611111111111111
|
|
GO:BP
|
GO:0045333
|
cellular respiration
|
2.29142526639134e-13
|
192
|
25
|
11
|
0.44
|
0.0572916666666667
|
|
GO:BP
|
GO:0008380
|
RNA splicing
|
3.04554359505924e-13
|
456
|
138
|
26
|
0.188405797101449
|
0.0570175438596491
|
|
GO:BP
|
GO:0015980
|
energy derivation by oxidation of organic compounds
|
1.2805525418623e-11
|
289
|
13
|
9
|
0.692307692307692
|
0.0311418685121107
|
|
GO:BP
|
GO:0006091
|
generation of precursor metabolites and energy
|
1.3105343913787e-11
|
538
|
25
|
13
|
0.52
|
0.0241635687732342
|
|
GO:BP
|
GO:0000398
|
mRNA splicing, via spliceosome
|
2.71123374208123e-11
|
361
|
129
|
21
|
0.162790697674419
|
0.0581717451523546
|
|
GO:BP
|
GO:0000377
|
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile
|
2.71123374208123e-11
|
361
|
129
|
21
|
0.162790697674419
|
0.0581717451523546
|
|
GO:BP
|
GO:0000375
|
RNA splicing, via transesterification reactions
|
2.9410386442011e-11
|
364
|
129
|
21
|
0.162790697674419
|
0.0576923076923077
|
|
GO:BP
|
GO:0006397
|
mRNA processing
|
5.34207115607952e-11
|
532
|
138
|
25
|
0.181159420289855
|
0.0469924812030075
|
|
GO:BP
|
GO:0006120
|
mitochondrial electron transport, NADH to ubiquinone
|
1.42791831529796e-10
|
56
|
25
|
7
|
0.28
|
0.125
|
|
GO:BP
|
GO:0015990
|
electron transport coupled proton transport
|
1.42791831529796e-10
|
5
|
13
|
4
|
0.307692307692308
|
0.8
|
|
GO:BP
|
GO:0015988
|
energy coupled proton transmembrane transport, against electrochemical gradient
|
1.42791831529796e-10
|
5
|
13
|
4
|
0.307692307692308
|
0.8
|
|
GO:BP
|
GO:0016071
|
mRNA metabolic process
|
1.48898571831085e-10
|
874
|
116
|
28
|
0.241379310344828
|
0.0320366132723112
|
|
GO:BP
|
GO:1902600
|
proton transmembrane transport
|
8.00289389944912e-10
|
157
|
13
|
7
|
0.538461538461538
|
0.0445859872611465
|
|
GO:BP
|
GO:0043484
|
regulation of RNA splicing
|
2.92341210618182e-09
|
160
|
138
|
14
|
0.101449275362319
|
0.0875
|
|
GO:BP
|
GO:0009060
|
aerobic respiration
|
1.9417231179819e-08
|
89
|
8
|
5
|
0.625
|
0.0561797752808989
|
|
GO:BP
|
GO:0032981
|
mitochondrial respiratory chain complex I assembly
|
2.45162726553606e-08
|
67
|
23
|
6
|
0.260869565217391
|
0.0895522388059701
|
|
GO:BP
|
GO:0010257
|
NADH dehydrogenase complex assembly
|
2.45162726553606e-08
|
67
|
23
|
6
|
0.260869565217391
|
0.0895522388059701
|
|
GO:BP
|
GO:0006396
|
RNA processing
|
8.71666802128329e-08
|
964
|
129
|
27
|
0.209302325581395
|
0.0280082987551867
|
|
GO:BP
|
GO:0033108
|
mitochondrial respiratory chain complex assembly
|
3.13908335381751e-07
|
103
|
23
|
6
|
0.260869565217391
|
0.058252427184466
|
|
GO:BP
|
GO:0006123
|
mitochondrial electron transport, cytochrome c to oxygen
|
4.60820804620474e-07
|
20
|
4
|
3
|
0.75
|
0.15
|
|
GO:BP
|
GO:0048024
|
regulation of mRNA splicing, via spliceosome
|
4.60820804620474e-07
|
119
|
116
|
10
|
0.0862068965517241
|
0.0840336134453782
|
|
GO:BP
|
GO:0016070
|
RNA metabolic process
|
4.60820804620474e-07
|
4710
|
117
|
60
|
0.512820512820513
|
0.0127388535031847
|
|
GO:BP
|
GO:0019646
|
aerobic electron transport chain
|
4.60820804620474e-07
|
20
|
4
|
3
|
0.75
|
0.15
|
|
GO:BP
|
GO:0045935
|
positive regulation of nucleobase-containing compound metabolic process
|
7.26619954185257e-07
|
1887
|
141
|
39
|
0.276595744680851
|
0.0206677265500795
|
|
GO:BP
|
GO:0055114
|
obsolete oxidation-reduction process
|
7.8573107665902e-07
|
745
|
13
|
8
|
0.615384615384615
|
0.010738255033557
|
|
GO:BP
|
GO:0051254
|
positive regulation of RNA metabolic process
|
2.03758762731203e-06
|
1714
|
141
|
36
|
0.25531914893617
|
0.0210035005834306
|
|
GO:BP
|
GO:0016310
|
phosphorylation
|
2.31070405371035e-06
|
2138
|
44
|
20
|
0.454545454545455
|
0.00935453695042095
|
|
GO:BP
|
GO:0050684
|
regulation of mRNA processing
|
5.99333161361333e-06
|
158
|
116
|
10
|
0.0862068965517241
|
0.0632911392405063
|
|
GO:BP
|
GO:0000380
|
alternative mRNA splicing, via spliceosome
|
7.3078301966086e-06
|
89
|
114
|
8
|
0.0701754385964912
|
0.0898876404494382
|
|
GO:BP
|
GO:0009628
|
response to abiotic stimulus
|
1.01906674138781e-05
|
1261
|
15
|
9
|
0.6
|
0.00713719270420301
|
|
GO:BP
|
GO:0090304
|
nucleic acid metabolic process
|
2.0170798183394e-05
|
5226
|
117
|
60
|
0.512820512820513
|
0.0114810562571757
|
|
GO:BP
|
GO:0098662
|
inorganic cation transmembrane transport
|
3.12708019631677e-05
|
794
|
13
|
7
|
0.538461538461538
|
0.00881612090680101
|
|
GO:BP
|
GO:0051252
|
regulation of RNA metabolic process
|
3.46336325661559e-05
|
3822
|
142
|
56
|
0.394366197183099
|
0.0146520146520147
|
|
GO:BP
|
GO:0006139
|
nucleobase-containing compound metabolic process
|
4.01185771264364e-05
|
5746
|
117
|
63
|
0.538461538461538
|
0.0109641489731987
|
|
GO:BP
|
GO:0010467
|
gene expression
|
4.09724411266063e-05
|
6179
|
117
|
66
|
0.564102564102564
|
0.0106813400226574
|
|
GO:BP
|
GO:0000381
|
regulation of alternative mRNA splicing, via spliceosome
|
4.09724411266063e-05
|
78
|
114
|
7
|
0.0614035087719298
|
0.0897435897435897
|
|
GO:BP
|
GO:0019219
|
regulation of nucleobase-containing compound metabolic process
|
5.09922148276162e-05
|
4091
|
104
|
46
|
0.442307692307692
|
0.0112441945734539
|
|
GO:BP
|
GO:0098660
|
inorganic ion transmembrane transport
|
5.54600865585889e-05
|
883
|
13
|
7
|
0.538461538461538
|
0.00792751981879955
|
|
GO:BP
|
GO:0098655
|
cation transmembrane transport
|
6.84553725929348e-05
|
914
|
13
|
7
|
0.538461538461538
|
0.00765864332603939
|
|
GO:BP
|
GO:0006979
|
response to oxidative stress
|
7.22815892935605e-05
|
466
|
15
|
6
|
0.4
|
0.0128755364806867
|
|
GO:BP
|
GO:0007005
|
mitochondrion organization
|
7.51176766250783e-05
|
561
|
13
|
6
|
0.461538461538462
|
0.0106951871657754
|
|
GO:BP
|
GO:0046483
|
heterocycle metabolic process
|
9.52865252291e-05
|
5904
|
117
|
63
|
0.538461538461538
|
0.0106707317073171
|
|
GO:BP
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
0.000101955458854937
|
3083
|
141
|
47
|
0.333333333333333
|
0.0152448913396043
|
|
GO:BP
|
GO:0045944
|
positive regulation of transcription by RNA polymerase II
|
0.000105809010355303
|
1197
|
108
|
22
|
0.203703703703704
|
0.0183792815371763
|
|
GO:BP
|
GO:0045893
|
positive regulation of transcription, DNA-templated
|
0.000118155314396156
|
1617
|
108
|
26
|
0.240740740740741
|
0.0160791589363018
|
|
GO:BP
|
GO:1903508
|
positive regulation of nucleic acid-templated transcription
|
0.000118155314396156
|
1617
|
108
|
26
|
0.240740740740741
|
0.0160791589363018
|
|
GO:BP
|
GO:1902680
|
positive regulation of RNA biosynthetic process
|
0.000118155314396156
|
1618
|
108
|
26
|
0.240740740740741
|
0.0160692212608158
|
|
GO:BP
|
GO:0006725
|
cellular aromatic compound metabolic process
|
0.000118155314396156
|
5956
|
117
|
63
|
0.538461538461538
|
0.0105775688381464
|
|
GO:BP
|
GO:0006796
|
phosphate-containing compound metabolic process
|
0.000126174236506103
|
3087
|
15
|
11
|
0.733333333333333
|
0.00356333009394234
|
|
GO:BP
|
GO:0006793
|
phosphorus metabolic process
|
0.00013552282908918
|
3114
|
15
|
11
|
0.733333333333333
|
0.00353243416827232
|
|
GO:BP
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
0.000138247880940615
|
6563
|
117
|
67
|
0.572649572649573
|
0.0102087460003047
|
|
GO:BP
|
GO:1903311
|
regulation of mRNA metabolic process
|
0.000150736106091976
|
356
|
116
|
12
|
0.103448275862069
|
0.0337078651685393
|
|
GO:BP
|
GO:0031325
|
positive regulation of cellular metabolic process
|
0.000192214898828519
|
3274
|
141
|
48
|
0.340425531914894
|
0.0146609651802077
|
|
GO:BP
|
GO:0070482
|
response to oxygen levels
|
0.000221130851387941
|
408
|
31
|
7
|
0.225806451612903
|
0.017156862745098
|
|
GO:BP
|
GO:2001014
|
regulation of skeletal muscle cell differentiation
|
0.000252590054283847
|
21
|
30
|
3
|
0.1
|
0.142857142857143
|
|
GO:BP
|
GO:0006812
|
cation transport
|
0.000302877791694606
|
1197
|
13
|
7
|
0.538461538461538
|
0.00584795321637427
|
|
GO:BP
|
GO:0097193
|
intrinsic apoptotic signaling pathway
|
0.000410124842588888
|
294
|
130
|
11
|
0.0846153846153846
|
0.0374149659863946
|
|
GO:BP
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
0.00043478388325715
|
3483
|
141
|
49
|
0.347517730496454
|
0.0140683318977893
|
|
GO:BP
|
GO:1901360
|
organic cyclic compound metabolic process
|
0.000436809298291528
|
6201
|
117
|
63
|
0.538461538461538
|
0.0101596516690856
|
|
GO:BP
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
0.000460563777441629
|
5848
|
104
|
55
|
0.528846153846154
|
0.00940492476060192
|
|
GO:BP
|
GO:1901796
|
regulation of signal transduction by p53 class mediator
|
0.000543328163048617
|
180
|
31
|
5
|
0.161290322580645
|
0.0277777777777778
|
|
GO:BP
|
GO:0030330
|
DNA damage response, signal transduction by p53 class mediator
|
0.000608920591314173
|
110
|
130
|
7
|
0.0538461538461538
|
0.0636363636363636
|
|
GO:BP
|
GO:0006325
|
chromatin organization
|
0.00074587933517193
|
824
|
104
|
16
|
0.153846153846154
|
0.0194174757281553
|
|
GO:BP
|
GO:0034220
|
ion transmembrane transport
|
0.00074587933517193
|
1397
|
13
|
7
|
0.538461538461538
|
0.00501073729420186
|
|
GO:BP
|
GO:0033044
|
regulation of chromosome organization
|
0.000761588593306909
|
279
|
97
|
9
|
0.0927835051546392
|
0.032258064516129
|
|
GO:BP
|
GO:0006950
|
response to stress
|
0.000774189452439768
|
4170
|
33
|
19
|
0.575757575757576
|
0.00455635491606715
|
|
GO:BP
|
GO:0009893
|
positive regulation of metabolic process
|
0.000774189452439768
|
3783
|
141
|
51
|
0.361702127659574
|
0.0134813639968279
|
|
GO:BP
|
GO:0009891
|
positive regulation of biosynthetic process
|
0.000853902559036798
|
2004
|
141
|
33
|
0.234042553191489
|
0.0164670658682635
|
|
GO:BP
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
0.000949622544387676
|
1851
|
108
|
26
|
0.240740740740741
|
0.0140464613722312
|
|
GO:BP
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
0.000949622544387676
|
1967
|
108
|
27
|
0.25
|
0.0137264870360956
|
|
GO:BP
|
GO:0072331
|
signal transduction by p53 class mediator
|
0.00095001459298855
|
268
|
59
|
7
|
0.11864406779661
|
0.0261194029850746
|
|
GO:BP
|
GO:0034968
|
histone lysine methylation
|
0.00107033481067187
|
113
|
141
|
7
|
0.049645390070922
|
0.0619469026548673
|
|
GO:BP
|
GO:0080090
|
regulation of primary metabolic process
|
0.00114051456774703
|
6053
|
104
|
55
|
0.528846153846154
|
0.00908640343631257
|
|
GO:BP
|
GO:0001666
|
response to hypoxia
|
0.00114051456774703
|
367
|
31
|
6
|
0.193548387096774
|
0.0163487738419619
|
|
GO:BP
|
GO:0018023
|
peptidyl-lysine trimethylation
|
0.00114051456774703
|
47
|
141
|
5
|
0.0354609929078014
|
0.106382978723404
|
|
GO:BP
|
GO:0000245
|
spliceosomal complex assembly
|
0.00121334683925325
|
59
|
114
|
5
|
0.043859649122807
|
0.0847457627118644
|
|
GO:BP
|
GO:0036293
|
response to decreased oxygen levels
|
0.00138373507451589
|
382
|
31
|
6
|
0.193548387096774
|
0.0157068062827225
|
|
GO:BP
|
GO:0010468
|
regulation of gene expression
|
0.00147857527420931
|
5196
|
117
|
54
|
0.461538461538462
|
0.0103926096997691
|
|
GO:BP
|
GO:0055085
|
transmembrane transport
|
0.00171968365149645
|
1635
|
13
|
7
|
0.538461538461538
|
0.00428134556574924
|
|
GO:BP
|
GO:0042770
|
signal transduction in response to DNA damage
|
0.0018061896037068
|
135
|
130
|
7
|
0.0538461538461538
|
0.0518518518518519
|
|
GO:BP
|
GO:0034622
|
cellular protein-containing complex assembly
|
0.0019395389818069
|
1107
|
114
|
19
|
0.166666666666667
|
0.017163504968383
|
|
GO:BP
|
GO:0061647
|
histone H3-K9 modification
|
0.00206112619004425
|
47
|
83
|
4
|
0.0481927710843374
|
0.0851063829787234
|
|
GO:BP
|
GO:0016570
|
histone modification
|
0.00225276804319055
|
464
|
141
|
13
|
0.0921985815602837
|
0.0280172413793103
|
|
GO:BP
|
GO:0018022
|
peptidyl-lysine methylation
|
0.00225749840405007
|
130
|
97
|
6
|
0.0618556701030928
|
0.0461538461538462
|
|
GO:BP
|
GO:2001251
|
negative regulation of chromosome organization
|
0.00237407577803005
|
92
|
86
|
5
|
0.0581395348837209
|
0.0543478260869565
|
|
GO:BP
|
GO:0008213
|
protein alkylation
|
0.00237407577803005
|
178
|
141
|
8
|
0.0567375886524823
|
0.0449438202247191
|
|
GO:BP
|
GO:0006479
|
protein methylation
|
0.00237407577803005
|
178
|
141
|
8
|
0.0567375886524823
|
0.0449438202247191
|
|
GO:BP
|
GO:0006366
|
transcription by RNA polymerase II
|
0.00246767841570147
|
2723
|
108
|
32
|
0.296296296296296
|
0.0117517443995593
|
|
GO:BP
|
GO:0051276
|
chromosome organization
|
0.00260872226673247
|
1258
|
104
|
19
|
0.182692307692308
|
0.0151033386327504
|
|
GO:BP
|
GO:0016569
|
covalent chromatin modification
|
0.00271521270312934
|
478
|
141
|
13
|
0.0921985815602837
|
0.0271966527196653
|
|
GO:BP
|
GO:0031323
|
regulation of cellular metabolic process
|
0.00271920040900641
|
6269
|
104
|
55
|
0.528846153846154
|
0.00877332907959802
|
|
GO:BP
|
GO:0009791
|
post-embryonic development
|
0.00273090317337186
|
84
|
99
|
5
|
0.0505050505050505
|
0.0595238095238095
|
|
GO:BP
|
GO:0016571
|
histone methylation
|
0.00274618786538043
|
138
|
97
|
6
|
0.0618556701030928
|
0.0434782608695652
|
|
GO:BP
|
GO:0031326
|
regulation of cellular biosynthetic process
|
0.00278184791048515
|
4273
|
104
|
42
|
0.403846153846154
|
0.00982915984086122
|
|
GO:BP
|
GO:0097190
|
apoptotic signaling pathway
|
0.00285514483372945
|
600
|
130
|
14
|
0.107692307692308
|
0.0233333333333333
|
|
GO:BP
|
GO:0065003
|
protein-containing complex assembly
|
0.00285514483372945
|
1680
|
114
|
24
|
0.210526315789474
|
0.0142857142857143
|
|
GO:BP
|
GO:0032870
|
cellular response to hormone stimulus
|
0.00325464039337822
|
645
|
108
|
13
|
0.12037037037037
|
0.0201550387596899
|
|
GO:BP
|
GO:0000956
|
nuclear-transcribed mRNA catabolic process
|
0.00333254891551005
|
214
|
94
|
7
|
0.074468085106383
|
0.0327102803738318
|
|
GO:BP
|
GO:0006357
|
regulation of transcription by RNA polymerase II
|
0.00393122989913207
|
2555
|
108
|
30
|
0.277777777777778
|
0.0117416829745597
|
|
GO:BP
|
GO:0003170
|
heart valve development
|
0.00395461359170787
|
63
|
30
|
3
|
0.1
|
0.0476190476190476
|
|
GO:BP
|
GO:0018027
|
peptidyl-lysine dimethylation
|
0.00395883026327786
|
23
|
83
|
3
|
0.036144578313253
|
0.130434782608696
|
|
GO:BP
|
GO:0045471
|
response to ethanol
|
0.00411236208098638
|
142
|
14
|
3
|
0.214285714285714
|
0.0211267605633803
|
|
GO:BP
|
GO:0009889
|
regulation of biosynthetic process
|
0.00411768406357886
|
4361
|
104
|
42
|
0.403846153846154
|
0.00963081861958266
|
|
GO:BP
|
GO:0006402
|
mRNA catabolic process
|
0.00425316816182102
|
384
|
94
|
9
|
0.0957446808510638
|
0.0234375
|
|
GO:BP
|
GO:0044237
|
cellular metabolic process
|
0.00425316816182102
|
10871
|
142
|
106
|
0.746478873239437
|
0.00975071290589642
|
|
GO:BP
|
GO:0033120
|
positive regulation of RNA splicing
|
0.00427436997682715
|
40
|
128
|
4
|
0.03125
|
0.1
|
|
GO:BP
|
GO:0031060
|
regulation of histone methylation
|
0.00427436997682715
|
67
|
141
|
5
|
0.0354609929078014
|
0.0746268656716418
|
|
GO:BP
|
GO:0034654
|
nucleobase-containing compound biosynthetic process
|
0.00427436997682715
|
4199
|
108
|
42
|
0.388888888888889
|
0.0100023815194094
|
|
GO:BP
|
GO:0035914
|
skeletal muscle cell differentiation
|
0.00451552886470606
|
68
|
30
|
3
|
0.1
|
0.0441176470588235
|
|
GO:BP
|
GO:1900542
|
regulation of purine nucleotide metabolic process
|
0.00492103761758617
|
119
|
18
|
3
|
0.166666666666667
|
0.0252100840336134
|
|
GO:BP
|
GO:0006140
|
regulation of nucleotide metabolic process
|
0.00512742253221158
|
121
|
18
|
3
|
0.166666666666667
|
0.0247933884297521
|
|
GO:BP
|
GO:0022618
|
ribonucleoprotein complex assembly
|
0.00570641092345748
|
197
|
114
|
7
|
0.0614035087719298
|
0.0355329949238579
|
|
GO:BP
|
GO:0051726
|
regulation of cell cycle
|
0.00570641092345748
|
1236
|
105
|
18
|
0.171428571428571
|
0.0145631067961165
|
|
GO:BP
|
GO:0031062
|
positive regulation of histone methylation
|
0.00587141807846299
|
40
|
141
|
4
|
0.0283687943262411
|
0.1
|
|
GO:BP
|
GO:0018130
|
heterocycle biosynthetic process
|
0.00591643728922681
|
4269
|
108
|
42
|
0.388888888888889
|
0.00983836964160225
|
|
GO:BP
|
GO:0006351
|
transcription, DNA-templated
|
0.00607307535005672
|
3714
|
108
|
38
|
0.351851851851852
|
0.0102315562735595
|
|
GO:BP
|
GO:0019438
|
aromatic compound biosynthetic process
|
0.00607307535005672
|
4278
|
108
|
42
|
0.388888888888889
|
0.00981767180925666
|
|
GO:BP
|
GO:0097659
|
nucleic acid-templated transcription
|
0.00607307535005672
|
3715
|
108
|
38
|
0.351851851851852
|
0.010228802153432
|
|
GO:BP
|
GO:0044271
|
cellular nitrogen compound biosynthetic process
|
0.00611819848315282
|
5017
|
105
|
46
|
0.438095238095238
|
0.00916882599162846
|
|
GO:BP
|
GO:2001252
|
positive regulation of chromosome organization
|
0.00611819848315282
|
168
|
97
|
6
|
0.0618556701030928
|
0.0357142857142857
|
|
GO:BP
|
GO:0007050
|
cell cycle arrest
|
0.00611819848315282
|
241
|
44
|
5
|
0.113636363636364
|
0.020746887966805
|
|
GO:BP
|
GO:0080182
|
histone H3-K4 trimethylation
|
0.00614508701781008
|
17
|
141
|
3
|
0.0212765957446809
|
0.176470588235294
|
|
GO:BP
|
GO:0032774
|
RNA biosynthetic process
|
0.00635621800456231
|
3731
|
108
|
38
|
0.351851851851852
|
0.0101849370142053
|
|
GO:BP
|
GO:0071826
|
ribonucleoprotein complex subunit organization
|
0.0064416801835132
|
204
|
114
|
7
|
0.0614035087719298
|
0.0343137254901961
|
|
GO:BP
|
GO:0032922
|
circadian regulation of gene expression
|
0.00654647160531989
|
67
|
88
|
4
|
0.0454545454545455
|
0.0597014925373134
|
|
GO:BP
|
GO:0051053
|
negative regulation of DNA metabolic process
|
0.0066031448217066
|
125
|
86
|
5
|
0.0581395348837209
|
0.04
|
|
GO:BP
|
GO:0018205
|
peptidyl-lysine modification
|
0.00675887591360298
|
405
|
141
|
11
|
0.0780141843971631
|
0.0271604938271605
|
|
GO:BP
|
GO:0045445
|
myoblast differentiation
|
0.00709560431267818
|
84
|
30
|
3
|
0.1
|
0.0357142857142857
|
|
GO:BP
|
GO:0006401
|
RNA catabolic process
|
0.00709560431267818
|
424
|
94
|
9
|
0.0957446808510638
|
0.0212264150943396
|
|
GO:BP
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
0.00709560431267818
|
4071
|
142
|
50
|
0.352112676056338
|
0.0122819945959224
|
|
GO:BP
|
GO:2001244
|
positive regulation of intrinsic apoptotic signaling pathway
|
0.00715469350864168
|
62
|
99
|
4
|
0.0404040404040404
|
0.0645161290322581
|
|
GO:BP
|
GO:0006355
|
regulation of transcription, DNA-templated
|
0.00715469350864168
|
3534
|
142
|
45
|
0.316901408450704
|
0.0127334465195246
|
|
GO:BP
|
GO:1903506
|
regulation of nucleic acid-templated transcription
|
0.00715469350864168
|
3535
|
142
|
45
|
0.316901408450704
|
0.0127298444130127
|
|
GO:BP
|
GO:2001141
|
regulation of RNA biosynthetic process
|
0.00719819704642672
|
3540
|
142
|
45
|
0.316901408450704
|
0.0127118644067797
|
|
GO:BP
|
GO:0006977
|
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest
|
0.00719819704642672
|
58
|
44
|
3
|
0.0681818181818182
|
0.0517241379310345
|
|
GO:BP
|
GO:0043984
|
histone H4-K16 acetylation
|
0.00725843629677545
|
20
|
131
|
3
|
0.0229007633587786
|
0.15
|
|
GO:BP
|
GO:1902400
|
intracellular signal transduction involved in G1 DNA damage checkpoint
|
0.00731256718228087
|
59
|
44
|
3
|
0.0681818181818182
|
0.0508474576271186
|
|
GO:BP
|
GO:0009152
|
purine ribonucleotide biosynthetic process
|
0.00731256718228087
|
179
|
15
|
3
|
0.2
|
0.0167597765363128
|
|
GO:BP
|
GO:0072431
|
signal transduction involved in mitotic G1 DNA damage checkpoint
|
0.00731256718228087
|
59
|
44
|
3
|
0.0681818181818182
|
0.0508474576271186
|
|
GO:BP
|
GO:2000637
|
positive regulation of gene silencing by miRNA
|
0.00741172986067756
|
28
|
94
|
3
|
0.0319148936170213
|
0.107142857142857
|
|
GO:BP
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
0.00775646011776881
|
4102
|
142
|
50
|
0.352112676056338
|
0.0121891760117016
|
|
GO:BP
|
GO:1902403
|
signal transduction involved in mitotic DNA integrity checkpoint
|
0.00781221838345111
|
61
|
44
|
3
|
0.0681818181818182
|
0.0491803278688525
|
|
GO:BP
|
GO:1902402
|
signal transduction involved in mitotic DNA damage checkpoint
|
0.00781221838345111
|
61
|
44
|
3
|
0.0681818181818182
|
0.0491803278688525
|
|
GO:BP
|
GO:0060148
|
positive regulation of posttranscriptional gene silencing
|
0.00798174989113539
|
29
|
94
|
3
|
0.0319148936170213
|
0.103448275862069
|
|
GO:BP
|
GO:0072413
|
signal transduction involved in mitotic cell cycle checkpoint
|
0.00843365374884902
|
63
|
44
|
3
|
0.0681818181818182
|
0.0476190476190476
|
|
GO:BP
|
GO:0009260
|
ribonucleotide biosynthetic process
|
0.00848674734251229
|
192
|
15
|
3
|
0.2
|
0.015625
|
|
GO:BP
|
GO:0031571
|
mitotic G1 DNA damage checkpoint
|
0.00917111801878186
|
66
|
44
|
3
|
0.0681818181818182
|
0.0454545454545455
|
|
GO:BP
|
GO:0046390
|
ribose phosphate biosynthetic process
|
0.00917111801878186
|
199
|
15
|
3
|
0.2
|
0.0150753768844221
|
|
GO:BP
|
GO:0000819
|
sister chromatid segregation
|
0.00928696620204115
|
193
|
96
|
6
|
0.0625
|
0.0310880829015544
|
|
GO:BP
|
GO:0006164
|
purine nucleotide biosynthetic process
|
0.00928696620204115
|
202
|
15
|
3
|
0.2
|
0.0148514851485149
|
|
GO:BP
|
GO:0044819
|
mitotic G1/S transition checkpoint
|
0.00931045819179425
|
67
|
44
|
3
|
0.0681818181818182
|
0.0447761194029851
|
|
GO:BP
|
GO:0044783
|
G1 DNA damage checkpoint
|
0.00931045819179425
|
67
|
44
|
3
|
0.0681818181818182
|
0.0447761194029851
|
|
GO:BP
|
GO:0009266
|
response to temperature stimulus
|
0.00948767878981586
|
228
|
31
|
4
|
0.129032258064516
|
0.0175438596491228
|
|
GO:BP
|
GO:0051567
|
histone H3-K9 methylation
|
0.00951366564053447
|
36
|
83
|
3
|
0.036144578313253
|
0.0833333333333333
|
|
GO:BP
|
GO:0010628
|
positive regulation of gene expression
|
0.0097818442948089
|
1160
|
130
|
19
|
0.146153846153846
|
0.0163793103448276
|
|
GO:BP
|
GO:0032204
|
regulation of telomere maintenance
|
0.0098983515766772
|
82
|
86
|
4
|
0.0465116279069767
|
0.0487804878048781
|
|
GO:BP
|
GO:1901362
|
organic cyclic compound biosynthetic process
|
0.00991760810394601
|
4439
|
108
|
42
|
0.388888888888889
|
0.00946159044829917
|
|
GO:BP
|
GO:0051236
|
establishment of RNA localization
|
0.00991792168047979
|
204
|
129
|
7
|
0.0542635658914729
|
0.0343137254901961
|
|
GO:BP
|
GO:0009150
|
purine ribonucleotide metabolic process
|
0.00991792168047979
|
414
|
18
|
4
|
0.222222222222222
|
0.00966183574879227
|
|
GO:BP
|
GO:0031100
|
animal organ regeneration
|
0.00991792168047979
|
77
|
92
|
4
|
0.0434782608695652
|
0.051948051948052
|
|
GO:BP
|
GO:0007568
|
aging
|
0.00991792168047979
|
329
|
10
|
3
|
0.3
|
0.00911854103343465
|
|
GO:BP
|
GO:1902275
|
regulation of chromatin organization
|
0.00991792168047979
|
196
|
97
|
6
|
0.0618556701030928
|
0.0306122448979592
|
|
GO:BP
|
GO:0072522
|
purine-containing compound biosynthetic process
|
0.00997331441312478
|
213
|
15
|
3
|
0.2
|
0.0140845070422535
|
|
GO:BP
|
GO:0042221
|
response to chemical
|
0.0105575074297818
|
4781
|
31
|
17
|
0.548387096774194
|
0.00355574147667852
|
|
GO:BP
|
GO:2001235
|
positive regulation of apoptotic signaling pathway
|
0.0105575074297818
|
129
|
99
|
5
|
0.0505050505050505
|
0.0387596899224806
|
|
GO:BP
|
GO:0090068
|
positive regulation of cell cycle process
|
0.0105575074297818
|
309
|
141
|
9
|
0.0638297872340425
|
0.029126213592233
|
|
GO:BP
|
GO:0032205
|
negative regulation of telomere maintenance
|
0.0105575074297818
|
37
|
86
|
3
|
0.0348837209302326
|
0.0810810810810811
|
|
GO:BP
|
GO:0031058
|
positive regulation of histone modification
|
0.0105901940817747
|
91
|
141
|
5
|
0.0354609929078014
|
0.0549450549450549
|
|
GO:BP
|
GO:0009259
|
ribonucleotide metabolic process
|
0.0108061081453623
|
431
|
18
|
4
|
0.222222222222222
|
0.00928074245939675
|
|
GO:BP
|
GO:0014070
|
response to organic cyclic compound
|
0.0108384564920024
|
967
|
38
|
8
|
0.210526315789474
|
0.00827300930713547
|
|
GO:BP
|
GO:2001022
|
positive regulation of response to DNA damage stimulus
|
0.0109211515699876
|
111
|
67
|
4
|
0.0597014925373134
|
0.036036036036036
|
|
GO:BP
|
GO:0072422
|
signal transduction involved in DNA damage checkpoint
|
0.011187499455478
|
75
|
44
|
3
|
0.0681818181818182
|
0.04
|
|
GO:BP
|
GO:0072401
|
signal transduction involved in DNA integrity checkpoint
|
0.011187499455478
|
75
|
44
|
3
|
0.0681818181818182
|
0.04
|
|
GO:BP
|
GO:0019693
|
ribose phosphate metabolic process
|
0.0112626074881319
|
441
|
18
|
4
|
0.222222222222222
|
0.0090702947845805
|
|
GO:BP
|
GO:0006996
|
organelle organization
|
0.0115060268588867
|
4104
|
107
|
39
|
0.364485981308411
|
0.00950292397660819
|
|
GO:BP
|
GO:0043933
|
protein-containing complex subunit organization
|
0.0116431415790887
|
1963
|
87
|
20
|
0.229885057471264
|
0.0101884870096791
|
|
GO:BP
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.0119328149423852
|
6603
|
104
|
54
|
0.519230769230769
|
0.00817810086324398
|
|
GO:BP
|
GO:1901700
|
response to oxygen-containing compound
|
0.0119328149423852
|
1739
|
64
|
15
|
0.234375
|
0.00862564692351926
|
|
GO:BP
|
GO:0006163
|
purine nucleotide metabolic process
|
0.0119328149423852
|
450
|
18
|
4
|
0.222222222222222
|
0.00888888888888889
|
|
GO:BP
|
GO:0097305
|
response to alcohol
|
0.0120716737622832
|
255
|
14
|
3
|
0.214285714285714
|
0.0117647058823529
|
|
GO:BP
|
GO:0072395
|
signal transduction involved in cell cycle checkpoint
|
0.0120716737622832
|
78
|
44
|
3
|
0.0681818181818182
|
0.0384615384615385
|
|
GO:BP
|
GO:0002931
|
response to ischemia
|
0.0125029660598766
|
58
|
60
|
3
|
0.05
|
0.0517241379310345
|
|
GO:BP
|
GO:0051568
|
histone H3-K4 methylation
|
0.0133020756035331
|
57
|
141
|
4
|
0.0283687943262411
|
0.0701754385964912
|
|
GO:BP
|
GO:0003231
|
cardiac ventricle development
|
0.0133020756035331
|
120
|
30
|
3
|
0.1
|
0.025
|
|
GO:BP
|
GO:0072521
|
purine-containing compound metabolic process
|
0.0137596954100502
|
473
|
18
|
4
|
0.222222222222222
|
0.00845665961945032
|
|
GO:BP
|
GO:1990845
|
adaptive thermogenesis
|
0.0139955187053918
|
155
|
91
|
5
|
0.0549450549450549
|
0.032258064516129
|
|
GO:BP
|
GO:0007623
|
circadian rhythm
|
0.0139955187053918
|
219
|
96
|
6
|
0.0625
|
0.0273972602739726
|
|
GO:BP
|
GO:0009725
|
response to hormone
|
0.01431463569175
|
921
|
108
|
14
|
0.12962962962963
|
0.0152008686210641
|
|
GO:BP
|
GO:0031056
|
regulation of histone modification
|
0.0143182319047972
|
150
|
141
|
6
|
0.0425531914893617
|
0.04
|
|
GO:BP
|
GO:0044085
|
cellular component biogenesis
|
0.0143641334539759
|
3257
|
14
|
8
|
0.571428571428571
|
0.00245624808105619
|
|
GO:BP
|
GO:0006376
|
mRNA splice site selection
|
0.0143641334539759
|
33
|
114
|
3
|
0.0263157894736842
|
0.0909090909090909
|
|
GO:BP
|
GO:0007093
|
mitotic cell cycle checkpoint
|
0.0146259635099478
|
164
|
130
|
6
|
0.0461538461538462
|
0.0365853658536585
|
|
GO:BP
|
GO:0033047
|
regulation of mitotic sister chromatid segregation
|
0.0146259635099478
|
45
|
84
|
3
|
0.0357142857142857
|
0.0666666666666667
|
|
GO:BP
|
GO:0045787
|
positive regulation of cell cycle
|
0.0149737406953535
|
403
|
141
|
10
|
0.0709219858156028
|
0.0248138957816377
|
|
GO:BP
|
GO:0048511
|
rhythmic process
|
0.0149737406953535
|
307
|
96
|
7
|
0.0729166666666667
|
0.0228013029315961
|
|
GO:BP
|
GO:0071158
|
positive regulation of cell cycle arrest
|
0.0153302124867687
|
88
|
44
|
3
|
0.0681818181818182
|
0.0340909090909091
|
|
GO:BP
|
GO:0009165
|
nucleotide biosynthetic process
|
0.0154475834932465
|
269
|
15
|
3
|
0.2
|
0.0111524163568773
|
|
GO:BP
|
GO:0060485
|
mesenchyme development
|
0.0154475834932465
|
291
|
30
|
4
|
0.133333333333333
|
0.013745704467354
|
|
GO:BP
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
0.0159191578241774
|
5051
|
105
|
44
|
0.419047619047619
|
0.00871114630766185
|
|
GO:BP
|
GO:1901293
|
nucleoside phosphate biosynthetic process
|
0.0159191578241774
|
273
|
15
|
3
|
0.2
|
0.010989010989011
|
|
GO:BP
|
GO:1905269
|
positive regulation of chromatin organization
|
0.0161469400268522
|
105
|
141
|
5
|
0.0354609929078014
|
0.0476190476190476
|
|
GO:BP
|
GO:0043967
|
histone H4 acetylation
|
0.0164248329924438
|
67
|
131
|
4
|
0.0305343511450382
|
0.0597014925373134
|
|
GO:BP
|
GO:0010629
|
negative regulation of gene expression
|
0.0165492997510312
|
1460
|
56
|
12
|
0.214285714285714
|
0.00821917808219178
|
|
GO:BP
|
GO:0120162
|
positive regulation of cold-induced thermogenesis
|
0.016680403530662
|
97
|
91
|
4
|
0.043956043956044
|
0.0412371134020619
|
|
GO:BP
|
GO:0044265
|
cellular macromolecule catabolic process
|
0.0171242912124456
|
1231
|
132
|
19
|
0.143939393939394
|
0.0154346060113729
|
|
GO:BP
|
GO:0006403
|
RNA localization
|
0.0180005126455184
|
238
|
129
|
7
|
0.0542635658914729
|
0.0294117647058824
|
|
GO:BP
|
GO:0071383
|
cellular response to steroid hormone stimulus
|
0.018261545727065
|
210
|
108
|
6
|
0.0555555555555556
|
0.0285714285714286
|
|
GO:BP
|
GO:0008631
|
intrinsic apoptotic signaling pathway in response to oxidative stress
|
0.0183134395885847
|
44
|
96
|
3
|
0.03125
|
0.0681818181818182
|
|
GO:BP
|
GO:0010035
|
response to inorganic substance
|
0.0190819978452723
|
575
|
41
|
6
|
0.146341463414634
|
0.0104347826086957
|
|
GO:BP
|
GO:0006974
|
cellular response to DNA damage stimulus
|
0.0191516304081864
|
888
|
117
|
14
|
0.11965811965812
|
0.0157657657657658
|
|
GO:BP
|
GO:0009117
|
nucleotide metabolic process
|
0.0191516304081864
|
544
|
18
|
4
|
0.222222222222222
|
0.00735294117647059
|
|
GO:BP
|
GO:0009059
|
macromolecule biosynthetic process
|
0.0191880802916407
|
5114
|
105
|
44
|
0.419047619047619
|
0.00860383261634728
|
|
GO:BP
|
GO:0006753
|
nucleoside phosphate metabolic process
|
0.0204562988336824
|
555
|
18
|
4
|
0.222222222222222
|
0.00720720720720721
|
|
GO:BP
|
GO:0010033
|
response to organic substance
|
0.0205500224546685
|
3412
|
67
|
23
|
0.343283582089552
|
0.00674091441969519
|
|
GO:BP
|
GO:0019222
|
regulation of metabolic process
|
0.0212460147506665
|
7148
|
104
|
56
|
0.538461538461538
|
0.00783435926133184
|
|
GO:BP
|
GO:0009057
|
macromolecule catabolic process
|
0.0224396193866333
|
1468
|
132
|
21
|
0.159090909090909
|
0.0143051771117166
|
|
GO:BP
|
GO:0032210
|
regulation of telomere maintenance via telomerase
|
0.0225662074398429
|
54
|
86
|
3
|
0.0348837209302326
|
0.0555555555555556
|
|
GO:BP
|
GO:0044773
|
mitotic DNA damage checkpoint
|
0.0226398261715357
|
106
|
44
|
3
|
0.0681818181818182
|
0.0283018867924528
|
|
GO:BP
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
0.0233969368945395
|
1707
|
130
|
23
|
0.176923076923077
|
0.0134739308728764
|
|
GO:BP
|
GO:0072332
|
intrinsic apoptotic signaling pathway by p53 class mediator
|
0.0233969368945395
|
77
|
130
|
4
|
0.0307692307692308
|
0.051948051948052
|
|
GO:BP
|
GO:1901698
|
response to nitrogen compound
|
0.0233969368945395
|
1172
|
102
|
15
|
0.147058823529412
|
0.0127986348122867
|
|
GO:BP
|
GO:0009636
|
response to toxic substance
|
0.0233969368945395
|
243
|
20
|
3
|
0.15
|
0.0123456790123457
|
|
GO:BP
|
GO:0007519
|
skeletal muscle tissue development
|
0.0234026250458299
|
160
|
30
|
3
|
0.1
|
0.01875
|
|
GO:BP
|
GO:0003205
|
cardiac chamber development
|
0.0237368768950409
|
161
|
30
|
3
|
0.1
|
0.0186335403726708
|
|
GO:BP
|
GO:0006915
|
apoptotic process
|
0.0240639785289175
|
1990
|
56
|
14
|
0.25
|
0.00703517587939699
|
|
GO:BP
|
GO:0009408
|
response to heat
|
0.0245130896265251
|
159
|
31
|
3
|
0.0967741935483871
|
0.0188679245283019
|
|
GO:BP
|
GO:0010564
|
regulation of cell cycle process
|
0.0245850008542122
|
809
|
105
|
12
|
0.114285714285714
|
0.0148331273176761
|
|
GO:BP
|
GO:1901699
|
cellular response to nitrogen compound
|
0.0246355851777571
|
724
|
102
|
11
|
0.107843137254902
|
0.0151933701657459
|
|
GO:BP
|
GO:0044774
|
mitotic DNA integrity checkpoint
|
0.0246889327153983
|
112
|
44
|
3
|
0.0681818181818182
|
0.0267857142857143
|
|
GO:BP
|
GO:0032259
|
methylation
|
0.0247846714059367
|
372
|
141
|
9
|
0.0638297872340425
|
0.0241935483870968
|
|
GO:BP
|
GO:0033554
|
cellular response to stress
|
0.0247846714059367
|
2111
|
59
|
15
|
0.254237288135593
|
0.00710563713879678
|
|
GO:BP
|
GO:0071407
|
cellular response to organic cyclic compound
|
0.0254762060365318
|
587
|
108
|
10
|
0.0925925925925926
|
0.0170357751277683
|
|
GO:BP
|
GO:0046777
|
protein autophosphorylation
|
0.0256899868025869
|
240
|
44
|
4
|
0.0909090909090909
|
0.0166666666666667
|
|
GO:BP
|
GO:0010243
|
response to organonitrogen compound
|
0.0256899868025869
|
1085
|
134
|
17
|
0.126865671641791
|
0.015668202764977
|
|
GO:BP
|
GO:0034655
|
nucleobase-containing compound catabolic process
|
0.0257473371012155
|
567
|
94
|
9
|
0.0957446808510638
|
0.0158730158730159
|
|
GO:BP
|
GO:0060538
|
skeletal muscle organ development
|
0.0257929085109553
|
170
|
30
|
3
|
0.1
|
0.0176470588235294
|
|
GO:BP
|
GO:0071156
|
regulation of cell cycle arrest
|
0.0257929085109553
|
115
|
44
|
3
|
0.0681818181818182
|
0.0260869565217391
|
|
GO:BP
|
GO:2000113
|
negative regulation of cellular macromolecule biosynthetic process
|
0.0259785626425473
|
1628
|
130
|
22
|
0.169230769230769
|
0.0135135135135135
|
|
GO:BP
|
GO:0071396
|
cellular response to lipid
|
0.025999135739364
|
578
|
31
|
5
|
0.161290322580645
|
0.00865051903114187
|
|
GO:BP
|
GO:0043523
|
regulation of neuron apoptotic process
|
0.0261073862225397
|
216
|
117
|
6
|
0.0512820512820513
|
0.0277777777777778
|
|
GO:BP
|
GO:0006306
|
DNA methylation
|
0.0266931977389274
|
76
|
141
|
4
|
0.0283687943262411
|
0.0526315789473684
|
|
GO:BP
|
GO:0009890
|
negative regulation of biosynthetic process
|
0.0266931977389274
|
1740
|
130
|
23
|
0.176923076923077
|
0.0132183908045977
|
|
GO:BP
|
GO:0006305
|
DNA alkylation
|
0.0266931977389274
|
76
|
141
|
4
|
0.0283687943262411
|
0.0526315789473684
|
|
GO:BP
|
GO:0055086
|
nucleobase-containing small molecule metabolic process
|
0.0271749809571542
|
629
|
18
|
4
|
0.222222222222222
|
0.00635930047694754
|
|
GO:BP
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
0.0272360307404647
|
1639
|
130
|
22
|
0.169230769230769
|
0.0134228187919463
|
|
GO:BP
|
GO:0043414
|
macromolecule methylation
|
0.0276913398448857
|
312
|
141
|
8
|
0.0567375886524823
|
0.0256410256410256
|
|
GO:BP
|
GO:0000122
|
negative regulation of transcription by RNA polymerase II
|
0.0280051216043903
|
901
|
30
|
6
|
0.2
|
0.00665926748057714
|
|
GO:BP
|
GO:0050657
|
nucleic acid transport
|
0.0280051216043903
|
201
|
129
|
6
|
0.0465116279069767
|
0.0298507462686567
|
|
GO:BP
|
GO:0007062
|
sister chromatid cohesion
|
0.0280051216043903
|
56
|
96
|
3
|
0.03125
|
0.0535714285714286
|
|
GO:BP
|
GO:0098813
|
nuclear chromosome segregation
|
0.0280051216043903
|
272
|
96
|
6
|
0.0625
|
0.0220588235294118
|
|
GO:BP
|
GO:0050658
|
RNA transport
|
0.0280051216043903
|
201
|
129
|
6
|
0.0465116279069767
|
0.0298507462686567
|
|
GO:BP
|
GO:0008219
|
cell death
|
0.028245309368683
|
2292
|
56
|
15
|
0.267857142857143
|
0.00654450261780105
|
|
GO:BP
|
GO:0045934
|
negative regulation of nucleobase-containing compound metabolic process
|
0.0282733814381934
|
1548
|
130
|
21
|
0.161538461538462
|
0.0135658914728682
|
|
GO:BP
|
GO:1904356
|
regulation of telomere maintenance via telomere lengthening
|
0.0283392394251528
|
63
|
86
|
3
|
0.0348837209302326
|
0.0476190476190476
|
|
GO:BP
|
GO:0007507
|
heart development
|
0.0303295361490877
|
578
|
33
|
5
|
0.151515151515152
|
0.00865051903114187
|
|
GO:BP
|
GO:0022402
|
cell cycle process
|
0.0303295361490877
|
1439
|
105
|
17
|
0.161904761904762
|
0.0118137595552467
|
|
GO:BP
|
GO:0006807
|
nitrogen compound metabolic process
|
0.0303295361490877
|
10078
|
132
|
89
|
0.674242424242424
|
0.00883111728517563
|
|
GO:BP
|
GO:2000134
|
negative regulation of G1/S transition of mitotic cell cycle
|
0.0303551135038517
|
128
|
44
|
3
|
0.0681818181818182
|
0.0234375
|
|
GO:BP
|
GO:0071385
|
cellular response to glucocorticoid stimulus
|
0.0304417984617055
|
57
|
99
|
3
|
0.0303030303030303
|
0.0526315789473684
|
|
GO:BP
|
GO:0010469
|
regulation of signaling receptor activity
|
0.0304417984617055
|
189
|
61
|
4
|
0.0655737704918033
|
0.0211640211640212
|
|
GO:BP
|
GO:0071495
|
cellular response to endogenous stimulus
|
0.0304417984617055
|
1422
|
64
|
12
|
0.1875
|
0.00843881856540084
|
|
GO:BP
|
GO:1902806
|
regulation of cell cycle G1/S phase transition
|
0.0304417984617055
|
206
|
56
|
4
|
0.0714285714285714
|
0.0194174757281553
|
|
GO:BP
|
GO:0022607
|
cellular component assembly
|
0.0322746825660535
|
3010
|
14
|
7
|
0.5
|
0.00232558139534884
|
|
GO:BP
|
GO:0046700
|
heterocycle catabolic process
|
0.0322746825660535
|
602
|
94
|
9
|
0.0957446808510638
|
0.0149501661129568
|
|
GO:BP
|
GO:0045652
|
regulation of megakaryocyte differentiation
|
0.0322746825660535
|
83
|
141
|
4
|
0.0283687943262411
|
0.0481927710843374
|
|
GO:BP
|
GO:1901137
|
carbohydrate derivative biosynthetic process
|
0.0326035934607207
|
686
|
18
|
4
|
0.222222222222222
|
0.00583090379008746
|
|
GO:BP
|
GO:0043525
|
positive regulation of neuron apoptotic process
|
0.0328505686087247
|
59
|
99
|
3
|
0.0303030303030303
|
0.0508474576271186
|
|
GO:BP
|
GO:1902807
|
negative regulation of cell cycle G1/S phase transition
|
0.0332650788359758
|
134
|
44
|
3
|
0.0681818181818182
|
0.0223880597014925
|
|
GO:BP
|
GO:0033045
|
regulation of sister chromatid segregation
|
0.0332650788359758
|
70
|
84
|
3
|
0.0357142857142857
|
0.0428571428571429
|
|
GO:BP
|
GO:0071417
|
cellular response to organonitrogen compound
|
0.0337558744685786
|
666
|
102
|
10
|
0.0980392156862745
|
0.015015015015015
|
|
GO:BP
|
GO:0044270
|
cellular nitrogen compound catabolic process
|
0.0337558744685786
|
608
|
94
|
9
|
0.0957446808510638
|
0.0148026315789474
|
|
GO:BP
|
GO:0007004
|
telomere maintenance via telomerase
|
0.0337558744685786
|
69
|
86
|
3
|
0.0348837209302326
|
0.0434782608695652
|
|
GO:BP
|
GO:0008152
|
metabolic process
|
0.0337558744685786
|
11827
|
117
|
90
|
0.769230769230769
|
0.00760970660353429
|
|
GO:BP
|
GO:0008630
|
intrinsic apoptotic signaling pathway in response to DNA damage
|
0.0341973420703747
|
103
|
117
|
4
|
0.0341880341880342
|
0.0388349514563107
|
|
GO:BP
|
GO:0045786
|
negative regulation of cell cycle
|
0.0342969821643963
|
647
|
44
|
6
|
0.136363636363636
|
0.00927357032457496
|
|
GO:BP
|
GO:0044249
|
cellular biosynthetic process
|
0.0346546647259955
|
6154
|
105
|
49
|
0.466666666666667
|
0.00796230094247644
|
|
GO:BP
|
GO:0000075
|
cell cycle checkpoint
|
0.034755298037548
|
216
|
130
|
6
|
0.0461538461538462
|
0.0277777777777778
|
|
GO:BP
|
GO:0016573
|
histone acetylation
|
0.034755298037548
|
157
|
78
|
4
|
0.0512820512820513
|
0.0254777070063694
|
|
GO:BP
|
GO:0006278
|
RNA-dependent DNA biosynthetic process
|
0.034755298037548
|
71
|
86
|
3
|
0.0348837209302326
|
0.0422535211267606
|
|
GO:BP
|
GO:0042542
|
response to hydrogen peroxide
|
0.034755298037548
|
150
|
41
|
3
|
0.0731707317073171
|
0.02
|
|
GO:BP
|
GO:0012501
|
programmed cell death
|
0.0347585589326457
|
2143
|
56
|
14
|
0.25
|
0.00653289780681288
|
|
GO:BP
|
GO:0071384
|
cellular response to corticosteroid stimulus
|
0.0355059863523195
|
63
|
99
|
3
|
0.0303030303030303
|
0.0476190476190476
|
|
GO:BP
|
GO:0019439
|
aromatic compound catabolic process
|
0.0359090216948326
|
623
|
94
|
9
|
0.0957446808510638
|
0.014446227929374
|
|
GO:BP
|
GO:0009058
|
biosynthetic process
|
0.0359559730176062
|
6340
|
105
|
50
|
0.476190476190476
|
0.00788643533123028
|
|
GO:BP
|
GO:0048863
|
stem cell differentiation
|
0.0359733811112883
|
258
|
78
|
5
|
0.0641025641025641
|
0.0193798449612403
|
|
GO:BP
|
GO:0051168
|
nuclear export
|
0.0359733811112883
|
204
|
140
|
6
|
0.0428571428571429
|
0.0294117647058824
|
|
GO:BP
|
GO:0018393
|
internal peptidyl-lysine acetylation
|
0.0367435751130134
|
162
|
78
|
4
|
0.0512820512820513
|
0.0246913580246914
|
|
GO:BP
|
GO:0062197
|
cellular response to chemical stress
|
0.0367451844369223
|
365
|
56
|
5
|
0.0892857142857143
|
0.0136986301369863
|
|
GO:BP
|
GO:0045892
|
negative regulation of transcription, DNA-templated
|
0.0367451844369223
|
1337
|
30
|
7
|
0.233333333333333
|
0.00523560209424084
|
|
GO:BP
|
GO:0036294
|
cellular response to decreased oxygen levels
|
0.0367451844369223
|
222
|
130
|
6
|
0.0461538461538462
|
0.027027027027027
|
|
GO:BP
|
GO:0018105
|
peptidyl-serine phosphorylation
|
0.0367451844369223
|
313
|
41
|
4
|
0.0975609756097561
|
0.012779552715655
|
|
GO:BP
|
GO:0009719
|
response to endogenous stimulus
|
0.0367451844369223
|
1683
|
64
|
13
|
0.203125
|
0.0077243018419489
|
|
GO:BP
|
GO:0001649
|
osteoblast differentiation
|
0.0367451844369223
|
232
|
55
|
4
|
0.0727272727272727
|
0.0172413793103448
|
|
GO:BP
|
GO:1903507
|
negative regulation of nucleic acid-templated transcription
|
0.0367451844369223
|
1339
|
30
|
7
|
0.233333333333333
|
0.00522778192681105
|
|
GO:BP
|
GO:1902679
|
negative regulation of RNA biosynthetic process
|
0.0367961481636633
|
1341
|
30
|
7
|
0.233333333333333
|
0.0052199850857569
|
|
GO:BP
|
GO:0010948
|
negative regulation of cell cycle process
|
0.0369204386941562
|
372
|
130
|
8
|
0.0615384615384615
|
0.021505376344086
|
|
GO:BP
|
GO:0006475
|
internal protein amino acid acetylation
|
0.0370172375578231
|
164
|
78
|
4
|
0.0512820512820513
|
0.024390243902439
|
|
GO:BP
|
GO:0051402
|
neuron apoptotic process
|
0.0382782159240982
|
250
|
117
|
6
|
0.0512820512820513
|
0.024
|
|
GO:BP
|
GO:0018193
|
peptidyl-amino acid modification
|
0.0394976755174059
|
1303
|
141
|
19
|
0.134751773049645
|
0.0145817344589409
|
|
GO:BP
|
GO:0106106
|
cold-induced thermogenesis
|
0.0395850799819876
|
144
|
91
|
4
|
0.043956043956044
|
0.0277777777777778
|
|
GO:BP
|
GO:0120161
|
regulation of cold-induced thermogenesis
|
0.0395850799819876
|
144
|
91
|
4
|
0.043956043956044
|
0.0277777777777778
|
|
GO:BP
|
GO:0043170
|
macromolecule metabolic process
|
0.0399582450229023
|
9796
|
142
|
92
|
0.647887323943662
|
0.00939158840342997
|
|
GO:BP
|
GO:0000077
|
DNA damage checkpoint
|
0.0402546831127111
|
154
|
44
|
3
|
0.0681818181818182
|
0.0194805194805195
|
|
GO:BP
|
GO:0018394
|
peptidyl-lysine acetylation
|
0.04169013037477
|
172
|
78
|
4
|
0.0512820512820513
|
0.0232558139534884
|
|
GO:BP
|
GO:1901576
|
organic substance biosynthetic process
|
0.04169013037477
|
6248
|
105
|
49
|
0.466666666666667
|
0.00784250960307298
|
|
GO:BP
|
GO:1901361
|
organic cyclic compound catabolic process
|
0.0422829994575396
|
651
|
94
|
9
|
0.0957446808510638
|
0.0138248847926267
|
|
GO:BP
|
GO:0048762
|
mesenchymal cell differentiation
|
0.0423392301240586
|
234
|
30
|
3
|
0.1
|
0.0128205128205128
|
|
GO:BP
|
GO:1901701
|
cellular response to oxygen-containing compound
|
0.0427217811825701
|
1227
|
130
|
17
|
0.130769230769231
|
0.0138549307253464
|
|
GO:BP
|
GO:0018209
|
peptidyl-serine modification
|
0.0432804997955567
|
336
|
41
|
4
|
0.0975609756097561
|
0.0119047619047619
|
|
GO:BP
|
GO:0010638
|
positive regulation of organelle organization
|
0.0436659122160552
|
603
|
102
|
9
|
0.0882352941176471
|
0.0149253731343284
|
|
GO:BP
|
GO:0010833
|
telomere maintenance via telomere lengthening
|
0.043698525286531
|
82
|
86
|
3
|
0.0348837209302326
|
0.0365853658536585
|
|
GO:BP
|
GO:0006811
|
ion transport
|
0.043698525286531
|
3661
|
5
|
4
|
0.8
|
0.00109259765091505
|
|
GO:BP
|
GO:0000070
|
mitotic sister chromatid segregation
|
0.0441125613298964
|
164
|
84
|
4
|
0.0476190476190476
|
0.024390243902439
|
|
GO:BP
|
GO:2001242
|
regulation of intrinsic apoptotic signaling pathway
|
0.0441125613298964
|
167
|
130
|
5
|
0.0384615384615385
|
0.029940119760479
|
|
GO:BP
|
GO:0140014
|
mitotic nuclear division
|
0.0442287034448606
|
302
|
102
|
6
|
0.0588235294117647
|
0.0198675496688742
|
|
GO:BP
|
GO:0022613
|
ribonucleoprotein complex biogenesis
|
0.0442287034448606
|
446
|
114
|
8
|
0.0701754385964912
|
0.0179372197309417
|
|
GO:BP
|
GO:0031570
|
DNA integrity checkpoint
|
0.0446399536613847
|
164
|
44
|
3
|
0.0681818181818182
|
0.0182926829268293
|
|
GO:BP
|
GO:0033043
|
regulation of organelle organization
|
0.0449296059094362
|
1203
|
102
|
14
|
0.137254901960784
|
0.0116375727348296
|
|
GO:BP
|
GO:0010942
|
positive regulation of cell death
|
0.0449635495784114
|
684
|
33
|
5
|
0.151515151515152
|
0.00730994152046784
|
|
GO:BP
|
GO:0051983
|
regulation of chromosome segregation
|
0.0449635495784114
|
86
|
84
|
3
|
0.0357142857142857
|
0.0348837209302326
|
|
GO:BP
|
GO:0051052
|
regulation of DNA metabolic process
|
0.045662102434806
|
363
|
86
|
6
|
0.0697674418604651
|
0.0165289256198347
|
|
GO:BP
|
GO:0044728
|
DNA methylation or demethylation
|
0.0460963886628015
|
100
|
141
|
4
|
0.0283687943262411
|
0.04
|
|
GO:BP
|
GO:0071453
|
cellular response to oxygen levels
|
0.0460963886628015
|
240
|
130
|
6
|
0.0461538461538462
|
0.025
|
|
GO:BP
|
GO:0030219
|
megakaryocyte differentiation
|
0.0460963886628015
|
100
|
141
|
4
|
0.0283687943262411
|
0.04
|
|
GO:BP
|
GO:0000723
|
telomere maintenance
|
0.0466911214250416
|
165
|
86
|
4
|
0.0465116279069767
|
0.0242424242424242
|
|
GO:BP
|
GO:0051253
|
negative regulation of RNA metabolic process
|
0.0474743236564762
|
1445
|
30
|
7
|
0.233333333333333
|
0.00484429065743945
|
|
GO:BP
|
GO:0000724
|
double-strand break repair via homologous recombination
|
0.0498428054866445
|
138
|
105
|
4
|
0.0380952380952381
|
0.0289855072463768
|
|
GO:CC
|
GO:0070469
|
respirasome
|
6.47375226242944e-17
|
103
|
25
|
11
|
0.44
|
0.106796116504854
|
|
GO:CC
|
GO:0098803
|
respiratory chain complex
|
6.47375226242944e-17
|
86
|
13
|
9
|
0.692307692307692
|
0.104651162790698
|
|
GO:CC
|
GO:1990204
|
oxidoreductase complex
|
8.92247954930948e-13
|
113
|
15
|
8
|
0.533333333333333
|
0.0707964601769911
|
|
GO:CC
|
GO:0098800
|
inner mitochondrial membrane protein complex
|
1.10408550515457e-12
|
146
|
13
|
8
|
0.615384615384615
|
0.0547945205479452
|
|
GO:CC
|
GO:0005743
|
mitochondrial inner membrane
|
2.93104745569779e-12
|
503
|
13
|
10
|
0.769230769230769
|
0.0198807157057654
|
|
GO:CC
|
GO:0005746
|
mitochondrial respirasome
|
3.84009284426698e-12
|
89
|
13
|
7
|
0.538461538461538
|
0.0786516853932584
|
|
GO:CC
|
GO:0098798
|
mitochondrial protein-containing complex
|
4.86879431385383e-12
|
270
|
15
|
9
|
0.6
|
0.0333333333333333
|
|
GO:CC
|
GO:0019866
|
organelle inner membrane
|
5.75913973826228e-12
|
564
|
13
|
10
|
0.769230769230769
|
0.0177304964539007
|
|
GO:CC
|
GO:0031966
|
mitochondrial membrane
|
5.19441315878586e-11
|
746
|
25
|
13
|
0.52
|
0.017426273458445
|
|
GO:CC
|
GO:0016604
|
nuclear body
|
5.19441315878586e-11
|
816
|
141
|
29
|
0.205673758865248
|
0.0355392156862745
|
|
GO:CC
|
GO:0005654
|
nucleoplasm
|
5.19441315878586e-11
|
4102
|
117
|
60
|
0.512820512820513
|
0.0146270112140419
|
|
GO:CC
|
GO:0031981
|
nuclear lumen
|
1.09284927734813e-10
|
4444
|
117
|
62
|
0.52991452991453
|
0.013951395139514
|
|
GO:CC
|
GO:0005740
|
mitochondrial envelope
|
1.09284927734813e-10
|
797
|
25
|
13
|
0.52
|
0.0163111668757842
|
|
GO:CC
|
GO:0016607
|
nuclear speck
|
7.50331806478068e-10
|
412
|
141
|
20
|
0.141843971631206
|
0.0485436893203883
|
|
GO:CC
|
GO:0045271
|
respiratory chain complex I
|
9.74285042725416e-10
|
51
|
25
|
6
|
0.24
|
0.117647058823529
|
|
GO:CC
|
GO:0030964
|
NADH dehydrogenase complex
|
9.74285042725416e-10
|
51
|
25
|
6
|
0.24
|
0.117647058823529
|
|
GO:CC
|
GO:0005747
|
mitochondrial respiratory chain complex I
|
9.74285042725416e-10
|
51
|
25
|
6
|
0.24
|
0.117647058823529
|
|
GO:CC
|
GO:0031975
|
envelope
|
5.89785292275294e-09
|
1237
|
13
|
10
|
0.769230769230769
|
0.00808407437348424
|
|
GO:CC
|
GO:0031967
|
organelle envelope
|
5.89785292275294e-09
|
1237
|
13
|
10
|
0.769230769230769
|
0.00808407437348424
|
|
GO:CC
|
GO:0098796
|
membrane protein complex
|
1.07746925953883e-08
|
1322
|
13
|
10
|
0.769230769230769
|
0.0075642965204236
|
|
GO:CC
|
GO:0043231
|
intracellular membrane-bounded organelle
|
1.5676831726421e-08
|
11296
|
117
|
100
|
0.854700854700855
|
0.00885269121813031
|
|
GO:CC
|
GO:0005739
|
mitochondrion
|
3.61989128802247e-08
|
1673
|
15
|
11
|
0.733333333333333
|
0.00657501494321578
|
|
GO:CC
|
GO:0070013
|
intracellular organelle lumen
|
3.61989128802247e-08
|
5402
|
117
|
64
|
0.547008547008547
|
0.0118474639022584
|
|
GO:CC
|
GO:0070069
|
cytochrome complex
|
4.08619657998871e-08
|
34
|
11
|
4
|
0.363636363636364
|
0.117647058823529
|
|
GO:CC
|
GO:0045277
|
respiratory chain complex IV
|
6.51548053502687e-08
|
21
|
4
|
3
|
0.75
|
0.142857142857143
|
|
GO:CC
|
GO:0043233
|
organelle lumen
|
1.07843093309121e-07
|
5565
|
117
|
64
|
0.547008547008547
|
0.0115004492362983
|
|
GO:CC
|
GO:0031974
|
membrane-enclosed lumen
|
1.07843093309121e-07
|
5565
|
117
|
64
|
0.547008547008547
|
0.0115004492362983
|
|
GO:CC
|
GO:0005634
|
nucleus
|
1.82750220404432e-07
|
7592
|
117
|
77
|
0.658119658119658
|
0.0101422550052687
|
|
GO:CC
|
GO:1902494
|
catalytic complex
|
1.50670343257442e-06
|
1441
|
26
|
12
|
0.461538461538462
|
0.00832755031228314
|
|
GO:CC
|
GO:0005681
|
spliceosomal complex
|
1.78416540120956e-06
|
191
|
138
|
11
|
0.0797101449275362
|
0.0575916230366492
|
|
GO:CC
|
GO:0043229
|
intracellular organelle
|
4.99633916920194e-06
|
12491
|
117
|
101
|
0.863247863247863
|
0.00808582179169002
|
|
GO:CC
|
GO:0140513
|
nuclear protein-containing complex
|
1.27339516277969e-05
|
1249
|
139
|
26
|
0.18705035971223
|
0.0208166533226581
|
|
GO:CC
|
GO:1990904
|
ribonucleoprotein complex
|
4.75516650423877e-05
|
688
|
116
|
16
|
0.137931034482759
|
0.0232558139534884
|
|
GO:CC
|
GO:0043227
|
membrane-bounded organelle
|
8.06826159652967e-05
|
12858
|
118
|
101
|
0.855932203389831
|
0.0078550318867631
|
|
GO:CC
|
GO:0032991
|
protein-containing complex
|
8.99002518116811e-05
|
5548
|
131
|
62
|
0.473282442748092
|
0.0111751982696467
|
|
GO:CC
|
GO:0031090
|
organelle membrane
|
9.76450982685923e-05
|
3612
|
13
|
10
|
0.769230769230769
|
0.00276854928017719
|
|
GO:CC
|
GO:0071013
|
catalytic step 2 spliceosome
|
0.000109988218304429
|
85
|
113
|
6
|
0.0530973451327434
|
0.0705882352941176
|
|
GO:CC
|
GO:0005622
|
intracellular anatomical structure
|
0.00239345935592491
|
14790
|
117
|
106
|
0.905982905982906
|
0.00716700473292765
|
|
GO:CC
|
GO:0036464
|
cytoplasmic ribonucleoprotein granule
|
0.00334627823282658
|
237
|
107
|
7
|
0.0654205607476635
|
0.029535864978903
|
|
GO:CC
|
GO:0035770
|
ribonucleoprotein granule
|
0.00428390140056091
|
248
|
107
|
7
|
0.0654205607476635
|
0.0282258064516129
|
|
GO:CC
|
GO:0016021
|
integral component of membrane
|
0.00481639763237806
|
5711
|
11
|
9
|
0.818181818181818
|
0.00157590614603397
|
|
GO:CC
|
GO:0043226
|
organelle
|
0.00548925113460906
|
14008
|
118
|
102
|
0.864406779661017
|
0.00728155339805825
|
|
GO:CC
|
GO:0031224
|
intrinsic component of membrane
|
0.00563154456687637
|
5868
|
11
|
9
|
0.818181818181818
|
0.00153374233128834
|
|
GO:CC
|
GO:0005801
|
cis-Golgi network
|
0.0113599636439262
|
69
|
63
|
3
|
0.0476190476190476
|
0.0434782608695652
|
|
GO:CC
|
GO:0098687
|
chromosomal region
|
0.0216217677724032
|
354
|
131
|
8
|
0.0610687022900763
|
0.0225988700564972
|
|
GO:CC
|
GO:0071011
|
precatalytic spliceosome
|
0.0235924030967351
|
52
|
112
|
3
|
0.0267857142857143
|
0.0576923076923077
|
|
GO:CC
|
GO:0090575
|
RNA polymerase II transcription regulator complex
|
0.0307448461112013
|
183
|
117
|
5
|
0.0427350427350427
|
0.0273224043715847
|
|
GO:CC
|
GO:0005694
|
chromosome
|
0.0313835278483575
|
2071
|
108
|
21
|
0.194444444444444
|
0.0101400289715113
|
|
GO:CC
|
GO:0016020
|
membrane
|
0.0313835278483575
|
9838
|
21
|
17
|
0.80952380952381
|
0.00172799349461273
|
|
GO:CC
|
GO:0010494
|
cytoplasmic stress granule
|
0.0398516467514823
|
75
|
107
|
3
|
0.0280373831775701
|
0.04
|
|
GO:CC
|
GO:0000932
|
P-body
|
0.0449740711220096
|
91
|
94
|
3
|
0.0319148936170213
|
0.032967032967033
|
|
GO:CC
|
GO:0000779
|
condensed chromosome, centromeric region
|
0.0458111177959147
|
124
|
131
|
4
|
0.0305343511450382
|
0.032258064516129
|
|
GO:MF
|
GO:0009055
|
electron transfer activity
|
8.69160510761172e-15
|
148
|
25
|
11
|
0.44
|
0.0743243243243243
|
|
GO:MF
|
GO:0003954
|
NADH dehydrogenase activity
|
1.28075746544653e-11
|
46
|
25
|
7
|
0.28
|
0.152173913043478
|
|
GO:MF
|
GO:0003723
|
RNA binding
|
1.28075746544653e-11
|
1943
|
116
|
42
|
0.362068965517241
|
0.0216160576428204
|
|
GO:MF
|
GO:0008137
|
NADH dehydrogenase (ubiquinone) activity
|
1.28075746544653e-11
|
46
|
25
|
7
|
0.28
|
0.152173913043478
|
|
GO:MF
|
GO:0050136
|
NADH dehydrogenase (quinone) activity
|
1.28075746544653e-11
|
46
|
25
|
7
|
0.28
|
0.152173913043478
|
|
GO:MF
|
GO:0003955
|
NAD(P)H dehydrogenase (quinone) activity
|
1.70854549595399e-11
|
49
|
25
|
7
|
0.28
|
0.142857142857143
|
|
GO:MF
|
GO:0016655
|
oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor
|
7.36264879056527e-11
|
61
|
25
|
7
|
0.28
|
0.114754098360656
|
|
GO:MF
|
GO:0003676
|
nucleic acid binding
|
6.11542258988681e-10
|
4276
|
116
|
60
|
0.517241379310345
|
0.0140318054256314
|
|
GO:MF
|
GO:0016651
|
oxidoreductase activity, acting on NAD(P)H
|
3.29206834458884e-09
|
107
|
25
|
7
|
0.28
|
0.0654205607476635
|
|
GO:MF
|
GO:0016491
|
oxidoreductase activity
|
8.28163753184581e-09
|
771
|
13
|
9
|
0.692307692307692
|
0.0116731517509728
|
|
GO:MF
|
GO:0004129
|
cytochrome-c oxidase activity
|
3.34637379654053e-07
|
27
|
4
|
3
|
0.75
|
0.111111111111111
|
|
GO:MF
|
GO:0015002
|
heme-copper terminal oxidase activity
|
3.34637379654053e-07
|
27
|
4
|
3
|
0.75
|
0.111111111111111
|
|
GO:MF
|
GO:0016676
|
oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor
|
3.34637379654053e-07
|
27
|
4
|
3
|
0.75
|
0.111111111111111
|
|
GO:MF
|
GO:0016675
|
oxidoreductase activity, acting on a heme group of donors
|
3.480088830334e-07
|
28
|
4
|
3
|
0.75
|
0.107142857142857
|
|
GO:MF
|
GO:1901363
|
heterocyclic compound binding
|
6.37194880797767e-06
|
6228
|
117
|
66
|
0.564102564102564
|
0.010597302504817
|
|
GO:MF
|
GO:0097159
|
organic cyclic compound binding
|
9.6006115869805e-06
|
6309
|
117
|
66
|
0.564102564102564
|
0.0104612458392772
|
|
GO:MF
|
GO:0015078
|
proton transmembrane transporter activity
|
9.6006115869805e-06
|
125
|
10
|
4
|
0.4
|
0.032
|
|
GO:MF
|
GO:0008187
|
poly-pyrimidine tract binding
|
0.000147213322693958
|
29
|
80
|
4
|
0.05
|
0.137931034482759
|
|
GO:MF
|
GO:0003677
|
DNA binding
|
0.000147213322693958
|
2486
|
108
|
32
|
0.296296296296296
|
0.0128720836685438
|
|
GO:MF
|
GO:0018024
|
histone-lysine N-methyltransferase activity
|
0.00153651603716415
|
44
|
97
|
4
|
0.0412371134020619
|
0.0909090909090909
|
|
GO:MF
|
GO:0003712
|
transcription coregulator activity
|
0.00153651603716415
|
506
|
117
|
12
|
0.102564102564103
|
0.0237154150197628
|
|
GO:MF
|
GO:0003824
|
catalytic activity
|
0.00162811323767517
|
5896
|
26
|
18
|
0.692307692307692
|
0.00305291723202171
|
|
GO:MF
|
GO:0022890
|
inorganic cation transmembrane transporter activity
|
0.00216324990083119
|
591
|
4
|
3
|
0.75
|
0.0050761421319797
|
|
GO:MF
|
GO:0003729
|
mRNA binding
|
0.00236931370035383
|
552
|
114
|
12
|
0.105263157894737
|
0.0217391304347826
|
|
GO:MF
|
GO:0008324
|
cation transmembrane transporter activity
|
0.00249600474488336
|
639
|
4
|
3
|
0.75
|
0.00469483568075117
|
|
GO:MF
|
GO:0042054
|
histone methyltransferase activity
|
0.00264449484075723
|
54
|
97
|
4
|
0.0412371134020619
|
0.0740740740740741
|
|
GO:MF
|
GO:0008266
|
poly(U) RNA binding
|
0.00264449484075723
|
26
|
80
|
3
|
0.0375
|
0.115384615384615
|
|
GO:MF
|
GO:0008276
|
protein methyltransferase activity
|
0.00327563010509199
|
87
|
119
|
5
|
0.0420168067226891
|
0.0574712643678161
|
|
GO:MF
|
GO:0001098
|
basal transcription machinery binding
|
0.00382327364454142
|
69
|
87
|
4
|
0.0459770114942529
|
0.0579710144927536
|
|
GO:MF
|
GO:0001099
|
basal RNA polymerase II transcription machinery binding
|
0.00382327364454142
|
69
|
87
|
4
|
0.0459770114942529
|
0.0579710144927536
|
|
GO:MF
|
GO:0016279
|
protein-lysine N-methyltransferase activity
|
0.00382327364454142
|
62
|
97
|
4
|
0.0412371134020619
|
0.0645161290322581
|
|
GO:MF
|
GO:0016278
|
lysine N-methyltransferase activity
|
0.00394310954450378
|
63
|
97
|
4
|
0.0412371134020619
|
0.0634920634920635
|
|
GO:MF
|
GO:0015318
|
inorganic molecular entity transmembrane transporter activity
|
0.00403390330330866
|
831
|
4
|
3
|
0.75
|
0.0036101083032491
|
|
GO:MF
|
GO:0015075
|
ion transmembrane transporter activity
|
0.00561716727029224
|
956
|
4
|
3
|
0.75
|
0.00313807531380753
|
|
GO:MF
|
GO:0000993
|
RNA polymerase II complex binding
|
0.0061933328680034
|
50
|
61
|
3
|
0.0491803278688525
|
0.06
|
|
GO:MF
|
GO:1990841
|
promoter-specific chromatin binding
|
0.00671589469935864
|
58
|
55
|
3
|
0.0545454545454545
|
0.0517241379310345
|
|
GO:MF
|
GO:0003727
|
single-stranded RNA binding
|
0.00671589469935864
|
92
|
80
|
4
|
0.05
|
0.0434782608695652
|
|
GO:MF
|
GO:0022857
|
transmembrane transporter activity
|
0.00717421336535016
|
1075
|
4
|
3
|
0.75
|
0.0027906976744186
|
|
GO:MF
|
GO:0043175
|
RNA polymerase core enzyme binding
|
0.007241503907634
|
55
|
61
|
3
|
0.0491803278688525
|
0.0545454545454545
|
|
GO:MF
|
GO:0005215
|
transporter activity
|
0.00871722406868238
|
1185
|
4
|
3
|
0.75
|
0.00253164556962025
|
|
GO:MF
|
GO:0047485
|
protein N-terminus binding
|
0.00924454222146842
|
111
|
130
|
5
|
0.0384615384615385
|
0.045045045045045
|
|
GO:MF
|
GO:0036002
|
pre-mRNA binding
|
0.0114943968914382
|
36
|
114
|
3
|
0.0263157894736842
|
0.0833333333333333
|
|
GO:MF
|
GO:0004386
|
helicase activity
|
0.0132385112621778
|
162
|
98
|
5
|
0.0510204081632653
|
0.0308641975308642
|
|
GO:MF
|
GO:0070063
|
RNA polymerase binding
|
0.013343952464486
|
72
|
61
|
3
|
0.0491803278688525
|
0.0416666666666667
|
|
GO:MF
|
GO:0008170
|
N-methyltransferase activity
|
0.0139630384123753
|
100
|
97
|
4
|
0.0412371134020619
|
0.04
|
|
GO:MF
|
GO:0003713
|
transcription coactivator activity
|
0.0141587329530667
|
280
|
117
|
7
|
0.0598290598290598
|
0.025
|
|
GO:MF
|
GO:0043021
|
ribonucleoprotein complex binding
|
0.0192527779000798
|
137
|
130
|
5
|
0.0384615384615385
|
0.0364963503649635
|
|
GO:MF
|
GO:0008168
|
methyltransferase activity
|
0.0195587252975559
|
222
|
119
|
6
|
0.0504201680672269
|
0.027027027027027
|
|
GO:MF
|
GO:0008757
|
S-adenosylmethionine-dependent methyltransferase activity
|
0.0224671939482908
|
161
|
119
|
5
|
0.0420168067226891
|
0.031055900621118
|
|
GO:MF
|
GO:0016741
|
transferase activity, transferring one-carbon groups
|
0.0224671939482908
|
232
|
119
|
6
|
0.0504201680672269
|
0.0258620689655172
|
|
GO:MF
|
GO:0140110
|
transcription regulator activity
|
0.0254987021133991
|
1935
|
139
|
25
|
0.179856115107914
|
0.0129198966408269
|
|
GO:MF
|
GO:0003682
|
chromatin binding
|
0.0304626185517029
|
571
|
108
|
9
|
0.0833333333333333
|
0.0157618213660245
|
|
GO:MF
|
GO:0003714
|
transcription corepressor activity
|
0.0311639022876008
|
187
|
117
|
5
|
0.0427350427350427
|
0.0267379679144385
|
|
GO:MF
|
GO:0051018
|
protein kinase A binding
|
0.0331221694241699
|
52
|
140
|
3
|
0.0214285714285714
|
0.0576923076923077
|
|
GO:MF
|
GO:0008134
|
transcription factor binding
|
0.0350508128376298
|
676
|
95
|
9
|
0.0947368421052632
|
0.0133136094674556
|
|
GO:MF
|
GO:0042393
|
histone binding
|
0.0450401310199625
|
241
|
141
|
6
|
0.0425531914893617
|
0.024896265560166
|
|
HP
|
HP:0000576
|
Centrocecal scotoma
|
4.46522564412286e-19
|
12
|
25
|
10
|
0.4
|
0.833333333333333
|
|
HP
|
HP:0001427
|
Mitochondrial inheritance
|
4.46522564412286e-19
|
18
|
25
|
11
|
0.44
|
0.611111111111111
|
|
HP
|
HP:0001112
|
Leber optic atrophy
|
7.95383927310601e-18
|
10
|
25
|
9
|
0.36
|
0.9
|
|
HP
|
HP:0004309
|
Ventricular preexcitation
|
9.35013447586851e-18
|
23
|
25
|
11
|
0.44
|
0.478260869565217
|
|
HP
|
HP:0002572
|
Episodic vomiting
|
9.52273276786427e-18
|
32
|
13
|
10
|
0.769230769230769
|
0.3125
|
|
HP
|
HP:0000631
|
Retinal arterial tortuosity
|
1.86880626586699e-17
|
11
|
25
|
9
|
0.36
|
0.818181818181818
|
|
HP
|
HP:0007768
|
Central retinal vessel vascular tortuosity
|
1.86880626586699e-17
|
11
|
25
|
9
|
0.36
|
0.818181818181818
|
|
HP
|
HP:0200125
|
Mitochondrial respiratory chain defects
|
4.56357507815173e-17
|
18
|
25
|
10
|
0.4
|
0.555555555555556
|
|
HP
|
HP:0003200
|
Ragged-red muscle fibers
|
6.9173844936148e-17
|
40
|
13
|
10
|
0.769230769230769
|
0.25
|
|
HP
|
HP:0007763
|
Retinal telangiectasia
|
7.6838724007144e-17
|
19
|
25
|
10
|
0.4
|
0.526315789473684
|
|
HP
|
HP:0003737
|
Mitochondrial myopathy
|
1.58414493137831e-15
|
54
|
13
|
10
|
0.769230769230769
|
0.185185185185185
|
|
HP
|
HP:0003800
|
Muscle abnormality related to mitochondrial dysfunction
|
2.04438371927482e-14
|
69
|
13
|
10
|
0.769230769230769
|
0.144927536231884
|
|
HP
|
HP:0005116
|
Arterial tortuosity
|
2.98514550215045e-14
|
20
|
25
|
9
|
0.36
|
0.45
|
|
HP
|
HP:0000622
|
Blurred vision
|
3.43796157586651e-14
|
32
|
25
|
10
|
0.4
|
0.3125
|
|
HP
|
HP:0007924
|
Slow decrease in visual acuity
|
3.43796157586651e-14
|
32
|
25
|
10
|
0.4
|
0.3125
|
|
HP
|
HP:0012766
|
Widened cerebral subarachnoid space
|
3.99395458871542e-14
|
8
|
23
|
7
|
0.304347826086957
|
0.875
|
|
HP
|
HP:0004948
|
Vascular tortuosity
|
6.71700128557931e-14
|
22
|
25
|
9
|
0.36
|
0.409090909090909
|
|
HP
|
HP:0002401
|
Stroke-like episode
|
1.00150302122723e-13
|
15
|
23
|
8
|
0.347826086956522
|
0.533333333333333
|
|
HP
|
HP:0007327
|
Mixed demyelinating and axonal polyneuropathy
|
1.50877030619589e-13
|
9
|
23
|
7
|
0.304347826086957
|
0.777777777777778
|
|
HP
|
HP:0012841
|
Retinal vascular tortuosity
|
4.3590356131231e-13
|
41
|
25
|
10
|
0.4
|
0.24390243902439
|
|
HP
|
HP:0005157
|
Concentric hypertrophic cardiomyopathy
|
4.53604221482784e-13
|
10
|
23
|
7
|
0.304347826086957
|
0.7
|
|
HP
|
HP:0001716
|
Wolff-Parkinson-White syndrome
|
9.51016348768442e-13
|
19
|
23
|
8
|
0.347826086956522
|
0.421052631578947
|
|
HP
|
HP:0000603
|
Central scotoma
|
1.04455129877118e-12
|
45
|
25
|
10
|
0.4
|
0.222222222222222
|
|
HP
|
HP:0003572
|
Low plasma citrulline
|
1.04455129877118e-12
|
11
|
23
|
7
|
0.304347826086957
|
0.636363636363636
|
|
HP
|
HP:0100611
|
Multiple glomerular cysts
|
1.04455129877118e-12
|
11
|
23
|
7
|
0.304347826086957
|
0.636363636363636
|
|
HP
|
HP:0008316
|
Abnormal mitochondria in muscle tissue
|
1.37071223757858e-12
|
38
|
13
|
8
|
0.615384615384615
|
0.210526315789474
|
|
HP
|
HP:0025268
|
Stuttering
|
2.31397020870622e-12
|
12
|
23
|
7
|
0.304347826086957
|
0.583333333333333
|
|
HP
|
HP:0007141
|
Sensorimotor neuropathy
|
2.42018313708516e-12
|
72
|
13
|
9
|
0.692307692307692
|
0.125
|
|
HP
|
HP:0031546
|
Cardiac conduction abnormality
|
3.60941618298775e-12
|
123
|
13
|
10
|
0.769230769230769
|
0.0813008130081301
|
|
HP
|
HP:0002174
|
Postural tremor
|
3.96461584021239e-12
|
52
|
25
|
10
|
0.4
|
0.192307692307692
|
|
HP
|
HP:0007067
|
Distal peripheral sensory neuropathy
|
4.35303578233817e-12
|
13
|
23
|
7
|
0.304347826086957
|
0.538461538461538
|
|
HP
|
HP:0000114
|
Proximal tubulopathy
|
7.77887987421383e-12
|
48
|
13
|
8
|
0.615384615384615
|
0.166666666666667
|
|
HP
|
HP:0003648
|
Lacticaciduria
|
7.77887987421383e-12
|
14
|
23
|
7
|
0.304347826086957
|
0.5
|
|
HP
|
HP:0001345
|
Psychotic mentation
|
7.77887987421383e-12
|
14
|
23
|
7
|
0.304347826086957
|
0.5
|
|
HP
|
HP:0011965
|
Abnormal circulating citrulline concentration
|
7.77887987421383e-12
|
14
|
23
|
7
|
0.304347826086957
|
0.5
|
|
HP
|
HP:0001138
|
Optic neuropathy
|
8.88079841204824e-12
|
57
|
25
|
10
|
0.4
|
0.175438596491228
|
|
HP
|
HP:0003287
|
Abnormality of mitochondrial metabolism
|
9.94538641015141e-12
|
139
|
13
|
10
|
0.769230769230769
|
0.0719424460431655
|
|
HP
|
HP:0000816
|
Abnormality of Krebs cycle metabolism
|
1.28943836677192e-11
|
15
|
23
|
7
|
0.304347826086957
|
0.466666666666667
|
|
HP
|
HP:0100027
|
Recurrent pancreatitis
|
1.28943836677192e-11
|
15
|
23
|
7
|
0.304347826086957
|
0.466666666666667
|
|
HP
|
HP:0002490
|
Increased CSF lactate
|
1.49860188605994e-11
|
91
|
13
|
9
|
0.692307692307692
|
0.0989010989010989
|
|
HP
|
HP:0030085
|
Abnormal CSF lactate level
|
1.49860188605994e-11
|
91
|
13
|
9
|
0.692307692307692
|
0.0989010989010989
|
|
HP
|
HP:0012707
|
Elevated brain lactate level by MRS
|
3.35628061331368e-11
|
17
|
23
|
7
|
0.304347826086957
|
0.411764705882353
|
|
HP
|
HP:0004885
|
Episodic respiratory distress
|
3.35628061331368e-11
|
17
|
23
|
7
|
0.304347826086957
|
0.411764705882353
|
|
HP
|
HP:0007159
|
Fluctuations in consciousness
|
3.35628061331368e-11
|
17
|
23
|
7
|
0.304347826086957
|
0.411764705882353
|
|
HP
|
HP:0003481
|
Segmental peripheral demyelination/remyelination
|
3.35628061331368e-11
|
17
|
23
|
7
|
0.304347826086957
|
0.411764705882353
|
|
HP
|
HP:0012103
|
Abnormality of the mitochondrion
|
4.07268745163641e-11
|
163
|
13
|
10
|
0.769230769230769
|
0.0613496932515337
|
|
HP
|
HP:0025045
|
Abnormal brain lactate level by MRS
|
5.24193982348118e-11
|
18
|
23
|
7
|
0.304347826086957
|
0.388888888888889
|
|
HP
|
HP:0004308
|
Ventricular arrhythmia
|
5.29782462612115e-11
|
106
|
13
|
9
|
0.692307692307692
|
0.0849056603773585
|
|
HP
|
HP:0025454
|
Abnormal CSF metabolite level
|
7.31413531202086e-11
|
110
|
13
|
9
|
0.692307692307692
|
0.0818181818181818
|
|
HP
|
HP:0012429
|
Aplasia/Hypoplasia of the cerebral white matter
|
7.77734435099608e-11
|
19
|
23
|
7
|
0.304347826086957
|
0.368421052631579
|
|
HP
|
HP:0000575
|
Scotoma
|
9.8970602787409e-11
|
74
|
25
|
10
|
0.4
|
0.135135135135135
|
|
HP
|
HP:0031434
|
Abnormal speech prosody
|
1.14689462913833e-10
|
20
|
23
|
7
|
0.304347826086957
|
0.35
|
|
HP
|
HP:0000630
|
Abnormal retinal artery morphology
|
1.44617476016158e-10
|
52
|
25
|
9
|
0.36
|
0.173076923076923
|
|
HP
|
HP:0033109
|
Abnormal circulating non-proteinogenic amino acid concentration
|
3.39711426037828e-10
|
23
|
23
|
7
|
0.304347826086957
|
0.304347826086957
|
|
HP
|
HP:0004389
|
Intestinal pseudo-obstruction
|
3.39711426037828e-10
|
23
|
23
|
7
|
0.304347826086957
|
0.304347826086957
|
|
HP
|
HP:0004303
|
Abnormal muscle fiber morphology
|
3.4202762531902e-10
|
205
|
13
|
10
|
0.769230769230769
|
0.0487804878048781
|
|
HP
|
HP:0001271
|
Polyneuropathy
|
3.81708860353866e-10
|
58
|
25
|
9
|
0.36
|
0.155172413793103
|
|
HP
|
HP:0001045
|
Vitiligo
|
6.27697831628721e-10
|
25
|
23
|
7
|
0.304347826086957
|
0.28
|
|
HP
|
HP:0001644
|
Dilated cardiomyopathy
|
7.70413888437349e-10
|
145
|
13
|
9
|
0.692307692307692
|
0.0620689655172414
|
|
HP
|
HP:0000590
|
Progressive external ophthalmoplegia
|
8.14153338095215e-10
|
26
|
23
|
7
|
0.304347826086957
|
0.269230769230769
|
|
HP
|
HP:0002331
|
Recurrent paroxysmal headache
|
8.14153338095215e-10
|
26
|
23
|
7
|
0.304347826086957
|
0.269230769230769
|
|
HP
|
HP:3000036
|
Abnormality of head blood vessel
|
8.90064880668433e-10
|
64
|
25
|
9
|
0.36
|
0.140625
|
|
HP
|
HP:0000580
|
Pigmentary retinopathy
|
1.04430488411937e-09
|
151
|
13
|
9
|
0.692307692307692
|
0.0596026490066225
|
|
HP
|
HP:0002883
|
Hyperventilation
|
1.04430488411937e-09
|
27
|
23
|
7
|
0.304347826086957
|
0.259259259259259
|
|
HP
|
HP:0000095
|
Abnormal renal glomerulus morphology
|
1.50724902301952e-09
|
158
|
13
|
9
|
0.692307692307692
|
0.0569620253164557
|
|
HP
|
HP:0031263
|
Abnormal renal corpuscle morphology
|
1.50724902301952e-09
|
158
|
13
|
9
|
0.692307692307692
|
0.0569620253164557
|
|
HP
|
HP:0010794
|
Impaired visuospatial constructive cognition
|
1.73609042685931e-09
|
29
|
23
|
7
|
0.304347826086957
|
0.241379310344828
|
|
HP
|
HP:0003128
|
Lactic acidosis
|
1.73609042685931e-09
|
161
|
13
|
9
|
0.692307692307692
|
0.0559006211180124
|
|
HP
|
HP:0000529
|
Progressive visual loss
|
1.799652050355e-09
|
101
|
25
|
10
|
0.4
|
0.099009900990099
|
|
HP
|
HP:0000124
|
Renal tubular dysfunction
|
1.9750203070175e-09
|
101
|
13
|
8
|
0.615384615384615
|
0.0792079207920792
|
|
HP
|
HP:0002483
|
Bulbar signs
|
2.10811093890823e-09
|
30
|
23
|
7
|
0.304347826086957
|
0.233333333333333
|
|
HP
|
HP:0002045
|
Hypothermia
|
2.10811093890823e-09
|
30
|
23
|
7
|
0.304347826086957
|
0.233333333333333
|
|
HP
|
HP:0002151
|
Increased serum lactate
|
2.93372486330485e-09
|
172
|
13
|
9
|
0.692307692307692
|
0.0523255813953488
|
|
HP
|
HP:0002069
|
Bilateral tonic-clonic seizure
|
2.93372486330485e-09
|
261
|
13
|
10
|
0.769230769230769
|
0.0383141762452107
|
|
HP
|
HP:0002013
|
Vomiting
|
3.24534193728231e-09
|
264
|
13
|
10
|
0.769230769230769
|
0.0378787878787879
|
|
HP
|
HP:0012705
|
Abnormal metabolic brain imaging by MRS
|
3.28129980213601e-09
|
32
|
23
|
7
|
0.304347826086957
|
0.21875
|
|
HP
|
HP:0007302
|
Bipolar affective disorder
|
5.14395120725701e-09
|
34
|
23
|
7
|
0.304347826086957
|
0.205882352941176
|
|
HP
|
HP:0011035
|
Abnormal renal cortex morphology
|
5.81004025808972e-09
|
187
|
13
|
9
|
0.692307692307692
|
0.0481283422459893
|
|
HP
|
HP:0000597
|
Ophthalmoparesis
|
5.81004025808972e-09
|
281
|
13
|
10
|
0.769230769230769
|
0.0355871886120996
|
|
HP
|
HP:0002135
|
Basal ganglia calcification
|
6.09324182832828e-09
|
35
|
23
|
7
|
0.304347826086957
|
0.2
|
|
HP
|
HP:0100754
|
Mania
|
6.09324182832828e-09
|
35
|
23
|
7
|
0.304347826086957
|
0.2
|
|
HP
|
HP:0000408
|
Progressive sensorineural hearing impairment
|
7.44834418441401e-09
|
36
|
23
|
7
|
0.304347826086957
|
0.194444444444444
|
|
HP
|
HP:0001945
|
Fever
|
8.59059599220541e-09
|
294
|
13
|
10
|
0.769230769230769
|
0.0340136054421769
|
|
HP
|
HP:0000829
|
Hypoparathyroidism
|
8.8342682794196e-09
|
37
|
23
|
7
|
0.304347826086957
|
0.189189189189189
|
|
HP
|
HP:0003348
|
Hyperalaninemia
|
8.8342682794196e-09
|
37
|
23
|
7
|
0.304347826086957
|
0.189189189189189
|
|
HP
|
HP:0001123
|
Visual field defect
|
9.35968515097697e-09
|
199
|
13
|
9
|
0.692307692307692
|
0.0452261306532663
|
|
HP
|
HP:0002134
|
Abnormality of the basal ganglia
|
1.01252734976081e-08
|
201
|
13
|
9
|
0.692307692307692
|
0.0447761194029851
|
|
HP
|
HP:0010916
|
Abnormal circulating alanine concentration
|
1.03099522616816e-08
|
38
|
23
|
7
|
0.304347826086957
|
0.184210526315789
|
|
HP
|
HP:0010915
|
Abnormal circulating pyruvate family amino acid concentration
|
1.03099522616816e-08
|
38
|
23
|
7
|
0.304347826086957
|
0.184210526315789
|
|
HP
|
HP:0000572
|
Visual loss
|
1.27583412637918e-08
|
207
|
13
|
9
|
0.692307692307692
|
0.0434782608695652
|
|
HP
|
HP:0012575
|
Abnormal nephron morphology
|
1.31777867273942e-08
|
208
|
13
|
9
|
0.692307692307692
|
0.0432692307692308
|
|
HP
|
HP:0007183
|
Focal T2 hyperintense basal ganglia lesion
|
1.43174264093284e-08
|
40
|
13
|
6
|
0.461538461538462
|
0.15
|
|
HP
|
HP:0001009
|
Telangiectasia
|
1.56483287086885e-08
|
135
|
31
|
11
|
0.354838709677419
|
0.0814814814814815
|
|
HP
|
HP:0012377
|
Hemianopia
|
1.63928995503103e-08
|
41
|
13
|
6
|
0.461538461538462
|
0.146341463414634
|
|
HP
|
HP:0007108
|
Demyelinating peripheral neuropathy
|
1.88982569463206e-08
|
42
|
13
|
6
|
0.461538461538462
|
0.142857142857143
|
|
HP
|
HP:0003546
|
Exercise intolerance
|
2.03304132156951e-08
|
82
|
13
|
7
|
0.538461538461538
|
0.0853658536585366
|
|
HP
|
HP:0000751
|
Personality changes
|
2.14810342563171e-08
|
43
|
13
|
6
|
0.461538461538462
|
0.13953488372093
|
|
HP
|
HP:0001730
|
Progressive hearing impairment
|
2.45858180681319e-08
|
44
|
13
|
6
|
0.461538461538462
|
0.136363636363636
|
|
HP
|
HP:0001269
|
Hemiparesis
|
2.52491450698824e-08
|
85
|
13
|
7
|
0.538461538461538
|
0.0823529411764706
|
|
HP
|
HP:0002381
|
Aphasia
|
2.52491450698824e-08
|
71
|
23
|
8
|
0.347826086956522
|
0.112676056338028
|
|
HP
|
HP:0001941
|
Acidosis
|
2.58531443513698e-08
|
335
|
13
|
10
|
0.769230769230769
|
0.0298507462686567
|
|
HP
|
HP:0002922
|
Increased CSF protein
|
2.66956159559232e-08
|
45
|
13
|
6
|
0.461538461538462
|
0.133333333333333
|
|
HP
|
HP:0100651
|
Type I diabetes mellitus
|
2.66956159559232e-08
|
45
|
13
|
6
|
0.461538461538462
|
0.133333333333333
|
|
HP
|
HP:0000097
|
Focal segmental glomerulosclerosis
|
2.66956159559232e-08
|
45
|
13
|
6
|
0.461538461538462
|
0.133333333333333
|
|
HP
|
HP:0025456
|
Abnormal CSF protein level
|
3.41138183564315e-08
|
47
|
13
|
6
|
0.461538461538462
|
0.127659574468085
|
|
HP
|
HP:0033108
|
Abnormal circulating proteinogenic amino acid derivative concentration
|
3.41138183564315e-08
|
47
|
13
|
6
|
0.461538461538462
|
0.127659574468085
|
|
HP
|
HP:0011096
|
Peripheral demyelination
|
3.41138183564315e-08
|
47
|
13
|
6
|
0.461538461538462
|
0.127659574468085
|
|
HP
|
HP:0001639
|
Hypertrophic cardiomyopathy
|
4.65216834230459e-08
|
244
|
13
|
9
|
0.692307692307692
|
0.0368852459016393
|
|
HP
|
HP:0012751
|
Abnormal basal ganglia MRI signal intensity
|
4.93591019679555e-08
|
50
|
13
|
6
|
0.461538461538462
|
0.12
|
|
HP
|
HP:0004360
|
Abnormality of acid-base homeostasis
|
5.20919561216346e-08
|
363
|
13
|
10
|
0.769230769230769
|
0.0275482093663912
|
|
HP
|
HP:0002345
|
Action tremor
|
5.20919561216346e-08
|
160
|
13
|
8
|
0.615384615384615
|
0.05
|
|
HP
|
HP:0000726
|
Dementia
|
6.25202702053508e-08
|
164
|
13
|
8
|
0.615384615384615
|
0.0487804878048781
|
|
HP
|
HP:0001298
|
Encephalopathy
|
7.84195052134866e-08
|
260
|
13
|
9
|
0.692307692307692
|
0.0346153846153846
|
|
HP
|
HP:0000544
|
External ophthalmoplegia
|
8.55181851595928e-08
|
55
|
13
|
6
|
0.461538461538462
|
0.109090909090909
|
|
HP
|
HP:0002076
|
Migraine
|
9.88622786717167e-08
|
105
|
13
|
7
|
0.538461538461538
|
0.0666666666666667
|
|
HP
|
HP:0000819
|
Diabetes mellitus
|
1.14773199371869e-07
|
395
|
13
|
10
|
0.769230769230769
|
0.0253164556962025
|
|
HP
|
HP:0004370
|
Abnormality of temperature regulation
|
1.22600338728513e-07
|
398
|
13
|
10
|
0.769230769230769
|
0.0251256281407035
|
|
HP
|
HP:0001952
|
Glucose intolerance
|
1.4433418524266e-07
|
405
|
13
|
10
|
0.769230769230769
|
0.0246913580246914
|
|
HP
|
HP:0008619
|
Bilateral sensorineural hearing impairment
|
1.72092120904075e-07
|
62
|
13
|
6
|
0.461538461538462
|
0.0967741935483871
|
|
HP
|
HP:0011767
|
Abnormality of the parathyroid physiology
|
1.88367191566057e-07
|
63
|
13
|
6
|
0.461538461538462
|
0.0952380952380952
|
|
HP
|
HP:0003198
|
Myopathy
|
1.93574113367505e-07
|
290
|
13
|
9
|
0.692307692307692
|
0.0310344827586207
|
|
HP
|
HP:0030956
|
Abnormality of cardiovascular system electrophysiology
|
2.1772259953263e-07
|
424
|
13
|
10
|
0.769230769230769
|
0.0235849056603774
|
|
HP
|
HP:0410263
|
Brain imaging abnormality
|
2.1772259953263e-07
|
95
|
23
|
8
|
0.347826086956522
|
0.0842105263157895
|
|
HP
|
HP:0002017
|
Nausea and vomiting
|
3.24293108148756e-07
|
442
|
13
|
10
|
0.769230769230769
|
0.0226244343891403
|
|
HP
|
HP:0002579
|
Gastrointestinal dysmotility
|
3.71126500809508e-07
|
71
|
13
|
6
|
0.461538461538462
|
0.0845070422535211
|
|
HP
|
HP:0008046
|
Abnormal retinal vascular morphology
|
3.71126500809508e-07
|
208
|
13
|
8
|
0.615384615384615
|
0.0384615384615385
|
|
HP
|
HP:0030895
|
Abnormal gastrointestinal motility
|
3.71126500809508e-07
|
71
|
13
|
6
|
0.461538461538462
|
0.0845070422535211
|
|
HP
|
HP:0007703
|
Abnormality of retinal pigmentation
|
4.2580489454244e-07
|
319
|
13
|
9
|
0.692307692307692
|
0.0282131661442006
|
|
HP
|
HP:0000828
|
Abnormality of the parathyroid gland
|
5.06992167336186e-07
|
75
|
13
|
6
|
0.461538461538462
|
0.08
|
|
HP
|
HP:0000096
|
Glomerular sclerosis
|
5.06992167336186e-07
|
75
|
13
|
6
|
0.461538461538462
|
0.08
|
|
HP
|
HP:0001733
|
Pancreatitis
|
5.45507141387776e-07
|
76
|
13
|
6
|
0.461538461538462
|
0.0789473684210526
|
|
HP
|
HP:0005162
|
Abnormal left ventricular function
|
5.84945030792049e-07
|
77
|
13
|
6
|
0.461538461538462
|
0.0779220779220779
|
|
HP
|
HP:0001297
|
Stroke
|
5.84945030792049e-07
|
138
|
13
|
7
|
0.538461538461538
|
0.0507246376811594
|
|
HP
|
HP:0003477
|
Peripheral axonal neuropathy
|
6.74729112639941e-07
|
141
|
13
|
7
|
0.538461538461538
|
0.049645390070922
|
|
HP
|
HP:0000091
|
Abnormal renal tubule morphology
|
7.23669526500065e-07
|
80
|
13
|
6
|
0.461538461538462
|
0.075
|
|
HP
|
HP:0011017
|
Abnormal cellular physiology
|
7.33646175972488e-07
|
485
|
13
|
10
|
0.769230769230769
|
0.0206185567010309
|
|
HP
|
HP:0001712
|
Left ventricular hypertrophy
|
8.28704267869454e-07
|
82
|
13
|
6
|
0.461538461538462
|
0.0731707317073171
|
|
HP
|
HP:0002921
|
Abnormality of the cerebrospinal fluid
|
8.31599851021889e-07
|
492
|
13
|
10
|
0.769230769230769
|
0.0203252032520325
|
|
HP
|
HP:0032943
|
Abnormal urine pH
|
8.72951995786554e-07
|
83
|
13
|
6
|
0.461538461538462
|
0.072289156626506
|
|
HP
|
HP:0012072
|
Aciduria
|
8.72951995786554e-07
|
83
|
13
|
6
|
0.461538461538462
|
0.072289156626506
|
|
HP
|
HP:0003829
|
Incomplete penetrance
|
8.89527723033535e-07
|
150
|
25
|
9
|
0.36
|
0.06
|
|
HP
|
HP:0025354
|
Abnormal cellular phenotype
|
9.64535559314029e-07
|
501
|
13
|
10
|
0.769230769230769
|
0.0199600798403194
|
|
HP
|
HP:0002315
|
Headache
|
1.00682109919888e-06
|
240
|
13
|
8
|
0.615384615384615
|
0.0333333333333333
|
|
HP
|
HP:0012469
|
Infantile spasms
|
1.1293217220268e-06
|
87
|
13
|
6
|
0.461538461538462
|
0.0689655172413793
|
|
HP
|
HP:0012704
|
Widened subarachnoid space
|
1.20196378657597e-06
|
88
|
13
|
6
|
0.461538461538462
|
0.0681818181818182
|
|
HP
|
HP:0004372
|
Reduced consciousness/confusion
|
1.25353734849149e-06
|
366
|
13
|
9
|
0.692307692307692
|
0.0245901639344262
|
|
HP
|
HP:0030872
|
Abnormal cardiac ventricular function
|
1.45234647393066e-06
|
91
|
13
|
6
|
0.461538461538462
|
0.0659340659340659
|
|
HP
|
HP:0000648
|
Optic atrophy
|
1.6905580809194e-06
|
533
|
13
|
10
|
0.769230769230769
|
0.0187617260787993
|
|
HP
|
HP:0000519
|
Developmental cataract
|
1.6905580809194e-06
|
126
|
23
|
8
|
0.347826086956522
|
0.0634920634920635
|
|
HP
|
HP:0011675
|
Arrhythmia
|
1.73566611160026e-06
|
381
|
13
|
9
|
0.692307692307692
|
0.0236220472440945
|
|
HP
|
HP:0000112
|
Nephropathy
|
2.21160550081521e-06
|
98
|
13
|
6
|
0.461538461538462
|
0.0612244897959184
|
|
HP
|
HP:0003130
|
Abnormal peripheral myelination
|
2.33562204136952e-06
|
99
|
13
|
6
|
0.461538461538462
|
0.0606060606060606
|
|
HP
|
HP:0000764
|
Peripheral axonal degeneration
|
2.44852208861693e-06
|
173
|
13
|
7
|
0.538461538461538
|
0.0404624277456647
|
|
HP
|
HP:0001711
|
Abnormal left ventricle morphology
|
2.75899925270369e-06
|
102
|
13
|
6
|
0.461538461538462
|
0.0588235294117647
|
|
HP
|
HP:0000709
|
Psychosis
|
3.08056984170584e-06
|
104
|
13
|
6
|
0.461538461538462
|
0.0576923076923077
|
|
HP
|
HP:0003812
|
Phenotypic variability
|
3.17075144997927e-06
|
410
|
13
|
9
|
0.692307692307692
|
0.0219512195121951
|
|
HP
|
HP:0000763
|
Sensory neuropathy
|
3.38703986615184e-06
|
182
|
13
|
7
|
0.538461538461538
|
0.0384615384615385
|
|
HP
|
HP:0011014
|
Abnormal glucose homeostasis
|
3.84372464207569e-06
|
584
|
13
|
10
|
0.769230769230769
|
0.0171232876712329
|
|
HP
|
HP:0009830
|
Peripheral neuropathy
|
4.08030008465042e-06
|
588
|
13
|
10
|
0.769230769230769
|
0.0170068027210884
|
|
HP
|
HP:0010783
|
Erythema
|
4.90854261982113e-06
|
113
|
13
|
6
|
0.461538461538462
|
0.0530973451327434
|
|
HP
|
HP:0004374
|
Hemiplegia/hemiparesis
|
6.10661713675529e-06
|
199
|
13
|
7
|
0.538461538461538
|
0.0351758793969849
|
|
HP
|
HP:0001638
|
Cardiomyopathy
|
6.57041324447768e-06
|
463
|
10
|
8
|
0.8
|
0.0172786177105832
|
|
HP
|
HP:0001714
|
Ventricular hypertrophy
|
6.57041324447768e-06
|
119
|
13
|
6
|
0.461538461538462
|
0.0504201680672269
|
|
HP
|
HP:0001268
|
Mental deterioration
|
6.65899502620112e-06
|
311
|
13
|
8
|
0.615384615384615
|
0.0257234726688103
|
|
HP
|
HP:0003112
|
Abnormal circulating amino acid concentration
|
6.81601664810489e-06
|
120
|
13
|
6
|
0.461538461538462
|
0.05
|
|
HP
|
HP:0012703
|
Abnormal subarachnoid space morphology
|
7.47670032827268e-06
|
122
|
13
|
6
|
0.461538461538462
|
0.0491803278688525
|
|
HP
|
HP:0004890
|
Elevated pulmonary artery pressure
|
7.61987949700898e-06
|
123
|
13
|
6
|
0.461538461538462
|
0.0487804878048781
|
|
HP
|
HP:0002514
|
Cerebral calcification
|
7.61987949700898e-06
|
123
|
13
|
6
|
0.461538461538462
|
0.0487804878048781
|
|
HP
|
HP:0033578
|
Pre-capillary pulmonary hypertension
|
7.61987949700898e-06
|
123
|
13
|
6
|
0.461538461538462
|
0.0487804878048781
|
|
HP
|
HP:0002354
|
Memory impairment
|
7.61987949700898e-06
|
123
|
13
|
6
|
0.461538461538462
|
0.0487804878048781
|
|
HP
|
HP:0002092
|
Pulmonary arterial hypertension
|
7.61987949700898e-06
|
123
|
13
|
6
|
0.461538461538462
|
0.0487804878048781
|
|
HP
|
HP:0008047
|
Abnormality of the vasculature of the eye
|
7.73222923369636e-06
|
319
|
13
|
8
|
0.615384615384615
|
0.0250783699059561
|
|
HP
|
HP:0012795
|
Abnormality of the optic disc
|
8.24288685817495e-06
|
638
|
13
|
10
|
0.769230769230769
|
0.0156739811912226
|
|
HP
|
HP:0001637
|
Abnormal myocardium morphology
|
9.15316789792443e-06
|
487
|
10
|
8
|
0.8
|
0.0164271047227926
|
|
HP
|
HP:0012447
|
Abnormal myelination
|
1.06734842126897e-05
|
497
|
10
|
8
|
0.8
|
0.0160965794768612
|
|
HP
|
HP:0012091
|
Abnormality of pancreas physiology
|
1.34501139411261e-05
|
136
|
13
|
6
|
0.461538461538462
|
0.0441176470588235
|
|
HP
|
HP:0011276
|
Vascular skin abnormality
|
1.38575038427284e-05
|
493
|
13
|
9
|
0.692307692307692
|
0.0182555780933063
|
|
HP
|
HP:0011097
|
Epileptic spasm
|
1.44226179138208e-05
|
170
|
23
|
8
|
0.347826086956522
|
0.0470588235294118
|
|
HP
|
HP:0000044
|
Hypogonadotropic hypogonadism
|
1.70762569594563e-05
|
142
|
13
|
6
|
0.461538461538462
|
0.0422535211267606
|
|
HP
|
HP:0001399
|
Hepatic failure
|
1.7700827583283e-05
|
143
|
13
|
6
|
0.461538461538462
|
0.041958041958042
|
|
HP
|
HP:0002123
|
Generalized myoclonic seizure
|
1.99004551216045e-05
|
146
|
13
|
6
|
0.461538461538462
|
0.0410958904109589
|
|
HP
|
HP:0000602
|
Ophthalmoplegia
|
2.49396684535873e-05
|
249
|
13
|
7
|
0.538461538461538
|
0.0281124497991968
|
|
HP
|
HP:0001425
|
Heterogeneous
|
3.2383820544828e-05
|
259
|
13
|
7
|
0.538461538461538
|
0.027027027027027
|
|
HP
|
HP:0000587
|
Abnormality of the optic nerve
|
3.33606151221417e-05
|
743
|
13
|
10
|
0.769230769230769
|
0.0134589502018843
|
|
HP
|
HP:0030875
|
Abnormality of pulmonary circulation
|
3.33908028385915e-05
|
160
|
13
|
6
|
0.461538461538462
|
0.0375
|
|
HP
|
HP:0003110
|
Abnormality of urine homeostasis
|
3.63737719902293e-05
|
586
|
10
|
8
|
0.8
|
0.0136518771331058
|
|
HP
|
HP:0002098
|
Respiratory distress
|
3.81880955686172e-05
|
164
|
13
|
6
|
0.461538461538462
|
0.0365853658536585
|
|
HP
|
HP:0100659
|
Abnormality of the cerebral vasculature
|
4.07179004323634e-05
|
269
|
13
|
7
|
0.538461538461538
|
0.0260223048327138
|
|
HP
|
HP:0032794
|
Myoclonic seizure
|
4.20224185947565e-05
|
167
|
13
|
6
|
0.461538461538462
|
0.0359281437125748
|
|
HP
|
HP:0000093
|
Proteinuria
|
4.32898305899586e-05
|
168
|
13
|
6
|
0.461538461538462
|
0.0357142857142857
|
|
HP
|
HP:0005978
|
Type II diabetes mellitus
|
4.45869217742918e-05
|
169
|
13
|
6
|
0.461538461538462
|
0.0355029585798817
|
|
HP
|
HP:0011344
|
Severe global developmental delay
|
5.44013482272882e-05
|
175
|
13
|
6
|
0.461538461538462
|
0.0342857142857143
|
|
HP
|
HP:0001274
|
Agenesis of corpus callosum
|
5.71672448007523e-05
|
284
|
13
|
7
|
0.538461538461538
|
0.0246478873239437
|
|
HP
|
HP:0000407
|
Sensorineural hearing impairment
|
5.94817692438288e-05
|
794
|
13
|
10
|
0.769230769230769
|
0.0125944584382872
|
|
HP
|
HP:0002120
|
Cerebral cortical atrophy
|
6.0722403716163e-05
|
287
|
13
|
7
|
0.538461538461538
|
0.024390243902439
|
|
HP
|
HP:0020129
|
Abnormal urine protein level
|
6.19421882315086e-05
|
181
|
8
|
5
|
0.625
|
0.0276243093922652
|
|
HP
|
HP:0032677
|
Generalized-onset motor seizure
|
6.45588338430578e-05
|
181
|
13
|
6
|
0.461538461538462
|
0.0331491712707182
|
|
HP
|
HP:0001410
|
Decreased liver function
|
7.06958023705477e-05
|
184
|
13
|
6
|
0.461538461538462
|
0.0326086956521739
|
|
HP
|
HP:0004302
|
Functional motor deficit
|
7.48686028151489e-05
|
437
|
13
|
8
|
0.615384615384615
|
0.0183066361556064
|
|
HP
|
HP:0002066
|
Gait ataxia
|
7.8544911850158e-05
|
188
|
13
|
6
|
0.461538461538462
|
0.0319148936170213
|
|
HP
|
HP:0025155
|
Abnormality of hepatobiliary system physiology
|
7.8544911850158e-05
|
188
|
13
|
6
|
0.461538461538462
|
0.0319148936170213
|
|
HP
|
HP:0008947
|
Infantile muscular hypotonia
|
7.8544911850158e-05
|
188
|
13
|
6
|
0.461538461538462
|
0.0319148936170213
|
|
HP
|
HP:0000998
|
Hypertrichosis
|
8.00342647926642e-05
|
217
|
23
|
8
|
0.347826086956522
|
0.0368663594470046
|
|
HP
|
HP:0001635
|
Congestive heart failure
|
9.90990767003499e-05
|
196
|
13
|
6
|
0.461538461538462
|
0.0306122448979592
|
|
HP
|
HP:0000759
|
Abnormal peripheral nervous system morphology
|
0.000101440872556201
|
845
|
13
|
10
|
0.769230769230769
|
0.0118343195266272
|
|
HP
|
HP:0010766
|
Ectopic calcification
|
0.000103895442417973
|
203
|
8
|
5
|
0.625
|
0.0246305418719212
|
|
HP
|
HP:0010549
|
Weakness due to upper motor neuron dysfunction
|
0.000104435872822985
|
459
|
13
|
8
|
0.615384615384615
|
0.0174291938997821
|
|
HP
|
HP:0100704
|
Cerebral visual impairment
|
0.000112490241704472
|
201
|
13
|
6
|
0.461538461538462
|
0.0298507462686567
|
|
HP
|
HP:0001332
|
Dystonia
|
0.000132722383149256
|
475
|
13
|
8
|
0.615384615384615
|
0.0168421052631579
|
|
HP
|
HP:0001336
|
Myoclonus
|
0.000132722383149256
|
326
|
13
|
7
|
0.538461538461538
|
0.0214723926380368
|
|
HP
|
HP:0001251
|
Ataxia
|
0.000132722383149256
|
872
|
13
|
10
|
0.769230769230769
|
0.0114678899082569
|
|
HP
|
HP:0000479
|
Abnormal retinal morphology
|
0.000139226581173382
|
877
|
13
|
10
|
0.769230769230769
|
0.0114025085518814
|
|
HP
|
HP:0011389
|
Functional abnormality of the inner ear
|
0.000147748522976352
|
883
|
13
|
10
|
0.769230769230769
|
0.0113250283125708
|
|
HP
|
HP:0004354
|
Abnormal circulating carboxylic acid concentration
|
0.000157127792398875
|
214
|
13
|
6
|
0.461538461538462
|
0.0280373831775701
|
|
HP
|
HP:0000510
|
Rod-cone dystrophy
|
0.00016066381809036
|
215
|
13
|
6
|
0.461538461538462
|
0.027906976744186
|
|
HP
|
HP:0001508
|
Failure to thrive
|
0.000161957811469699
|
893
|
13
|
10
|
0.769230769230769
|
0.0111982082866741
|
|
HP
|
HP:0000359
|
Abnormality of the inner ear
|
0.000164633858245151
|
895
|
13
|
10
|
0.769230769230769
|
0.0111731843575419
|
|
HP
|
HP:0012210
|
Abnormal renal morphology
|
0.000189569500834752
|
909
|
13
|
10
|
0.769230769230769
|
0.011001100110011
|
|
HP
|
HP:0001328
|
Specific learning disability
|
0.000205422995796899
|
249
|
23
|
8
|
0.347826086956522
|
0.0321285140562249
|
|
HP
|
HP:0002793
|
Abnormal pattern of respiration
|
0.000209185241531332
|
352
|
13
|
7
|
0.538461538461538
|
0.0198863636363636
|
|
HP
|
HP:0100660
|
Dyskinesia
|
0.000209185241531332
|
226
|
13
|
6
|
0.461538461538462
|
0.0265486725663717
|
|
HP
|
HP:0001337
|
Tremor
|
0.000241944602023251
|
518
|
13
|
8
|
0.615384615384615
|
0.0154440154440154
|
|
HP
|
HP:0030163
|
Abnormal vascular physiology
|
0.000256765722944116
|
248
|
8
|
5
|
0.625
|
0.0201612903225806
|
|
HP
|
HP:0012211
|
Abnormal renal physiology
|
0.000257513333148821
|
614
|
9
|
7
|
0.777777777777778
|
0.011400651465798
|
|
HP
|
HP:0007359
|
Focal-onset seizure
|
0.000280600682202666
|
253
|
8
|
5
|
0.625
|
0.0197628458498024
|
|
HP
|
HP:0012748
|
Focal T2 hyperintense brainstem lesion
|
0.000281784379005803
|
32
|
9
|
3
|
0.333333333333333
|
0.09375
|
|
HP
|
HP:0002072
|
Chorea
|
0.000300416778215129
|
242
|
13
|
6
|
0.461538461538462
|
0.0247933884297521
|
|
HP
|
HP:0001732
|
Abnormality of the pancreas
|
0.000340949201079615
|
264
|
8
|
5
|
0.625
|
0.0189393939393939
|
|
HP
|
HP:0004305
|
Involuntary movements
|
0.000357485455475283
|
978
|
13
|
10
|
0.769230769230769
|
0.0102249488752556
|
|
HP
|
HP:0011443
|
Abnormality of coordination
|
0.000376777306774482
|
984
|
13
|
10
|
0.769230769230769
|
0.0101626016260163
|
|
HP
|
HP:0010993
|
Abnormality of the cerebral subcortex
|
0.000435824498314778
|
1000
|
13
|
10
|
0.769230769230769
|
0.01
|
|
HP
|
HP:0007704
|
Paroxysmal involuntary eye movements
|
0.00046405331953247
|
38
|
9
|
3
|
0.333333333333333
|
0.0789473684210526
|
|
HP
|
HP:0000821
|
Hypothyroidism
|
0.000468241710490391
|
283
|
8
|
5
|
0.625
|
0.0176678445229682
|
|
HP
|
HP:0012747
|
Abnormal brainstem MRI signal intensity
|
0.000497925710069945
|
39
|
9
|
3
|
0.333333333333333
|
0.0769230769230769
|
|
HP
|
HP:0000505
|
Visual impairment
|
0.000505523663866112
|
1018
|
13
|
10
|
0.769230769230769
|
0.00982318271119843
|
|
HP
|
HP:0007700
|
Ocular anterior segment dysgenesis
|
0.000509378567262534
|
285
|
23
|
8
|
0.347826086956522
|
0.0280701754385965
|
|
HP
|
HP:0020219
|
Motor seizure
|
0.000546985305153049
|
288
|
23
|
8
|
0.347826086956522
|
0.0277777777777778
|
|
HP
|
HP:0011004
|
Abnormal systemic arterial morphology
|
0.00056036715659321
|
435
|
25
|
10
|
0.4
|
0.0229885057471264
|
|
HP
|
HP:0002926
|
Abnormality of thyroid physiology
|
0.00056036715659321
|
295
|
8
|
5
|
0.625
|
0.0169491525423729
|
|
HP
|
HP:0025116
|
Fetal distress
|
0.000565135276062478
|
41
|
9
|
3
|
0.333333333333333
|
0.0731707317073171
|
|
HP
|
HP:0100543
|
Cognitive impairment
|
0.000640329047813833
|
597
|
13
|
8
|
0.615384615384615
|
0.0134003350083752
|
|
HP
|
HP:0002104
|
Apnea
|
0.000711736623604628
|
285
|
13
|
6
|
0.461538461538462
|
0.0210526315789474
|
|
HP
|
HP:0002014
|
Diarrhea
|
0.000711736623604628
|
311
|
8
|
5
|
0.625
|
0.0160771704180064
|
|
HP
|
HP:0001324
|
Muscle weakness
|
0.000742005513930796
|
898
|
10
|
8
|
0.8
|
0.0089086859688196
|
|
HP
|
HP:0000739
|
Anxiety
|
0.000784570889792254
|
318
|
8
|
5
|
0.625
|
0.0157232704402516
|
|
HP
|
HP:0003542
|
Increased serum pyruvate
|
0.000830231983123987
|
47
|
9
|
3
|
0.333333333333333
|
0.0638297872340425
|
|
HP
|
HP:0004366
|
Abnormality of glycolysis
|
0.000830231983123987
|
47
|
9
|
3
|
0.333333333333333
|
0.0638297872340425
|
|
HP
|
HP:0012719
|
Functional abnormality of the gastrointestinal tract
|
0.000855853401006664
|
917
|
10
|
8
|
0.8
|
0.00872410032715376
|
|
HP
|
HP:0003828
|
Variable expressivity
|
0.000876611936693102
|
297
|
13
|
6
|
0.461538461538462
|
0.0202020202020202
|
|
HP
|
HP:0011277
|
Abnormality of the urinary system physiology
|
0.000884764737454363
|
922
|
10
|
8
|
0.8
|
0.00867678958785249
|
|
HP
|
HP:0000716
|
Depressivity
|
0.000889640156867775
|
328
|
8
|
5
|
0.625
|
0.0152439024390244
|
|
HP
|
HP:0011923
|
Decreased activity of mitochondrial complex I
|
0.000922755818345538
|
49
|
9
|
3
|
0.333333333333333
|
0.0612244897959184
|
|
HP
|
HP:0002019
|
Constipation
|
0.000935885714352038
|
332
|
8
|
5
|
0.625
|
0.0150602409638554
|
|
HP
|
HP:0002197
|
Generalized-onset seizure
|
0.000947232349463085
|
314
|
23
|
8
|
0.347826086956522
|
0.0254777070063694
|
|
HP
|
HP:0100852
|
Abnormal fear/anxiety-related behavior
|
0.000998098337273042
|
337
|
8
|
5
|
0.625
|
0.0148367952522255
|
|
HP
|
HP:0000822
|
Hypertension
|
0.00100857230207036
|
306
|
13
|
6
|
0.461538461538462
|
0.0196078431372549
|
|
HP
|
HP:0002012
|
Abnormality of the abdominal organs
|
0.00103523819970854
|
1111
|
13
|
10
|
0.769230769230769
|
0.009000900090009
|
|
HP
|
HP:0011354
|
Generalized abnormality of skin
|
0.0012162641945033
|
876
|
13
|
9
|
0.692307692307692
|
0.0102739726027397
|
|
HP
|
HP:0002376
|
Developmental regression
|
0.00125885998866422
|
319
|
13
|
6
|
0.461538461538462
|
0.0188087774294671
|
|
HP
|
HP:0011025
|
Abnormal cardiovascular system physiology
|
0.00135446159945789
|
1147
|
11
|
9
|
0.818181818181818
|
0.00784655623365301
|
|
HP
|
HP:0000543
|
Optic disc pallor
|
0.00145300606039327
|
139
|
10
|
4
|
0.4
|
0.0287769784172662
|
|
HP
|
HP:0001288
|
Gait disturbance
|
0.00151362439331425
|
996
|
10
|
8
|
0.8
|
0.00803212851405622
|
|
HP
|
HP:0000556
|
Retinal dystrophy
|
0.00152379911473145
|
331
|
13
|
6
|
0.461538461538462
|
0.0181268882175227
|
|
HP
|
HP:0002240
|
Hepatomegaly
|
0.00155522577178852
|
492
|
13
|
7
|
0.538461538461538
|
0.0142276422764228
|
|
HP
|
HP:0011013
|
Abnormal circulating carbohydrate concentration
|
0.00161181723534252
|
60
|
9
|
3
|
0.333333333333333
|
0.05
|
|
HP
|
HP:0000818
|
Abnormality of the endocrine system
|
0.00166307939395719
|
1179
|
11
|
9
|
0.818181818181818
|
0.00763358778625954
|
|
HP
|
HP:0000135
|
Hypogonadism
|
0.00166307939395719
|
378
|
8
|
5
|
0.625
|
0.0132275132275132
|
|
HP
|
HP:0000077
|
Abnormality of the kidney
|
0.00168066945039127
|
1181
|
11
|
9
|
0.818181818181818
|
0.00762066045723963
|
|
HP
|
HP:0002493
|
Upper motor neuron dysfunction
|
0.00168638575749752
|
1182
|
11
|
9
|
0.818181818181818
|
0.00761421319796954
|
|
HP
|
HP:0001098
|
Abnormal fundus morphology
|
0.00179079377368628
|
1191
|
11
|
9
|
0.818181818181818
|
0.00755667506297229
|
|
HP
|
HP:0004329
|
Abnormal posterior eye segment morphology
|
0.00182231003302657
|
1194
|
11
|
9
|
0.818181818181818
|
0.00753768844221105
|
|
HP
|
HP:0000820
|
Abnormality of the thyroid gland
|
0.00184369641337984
|
388
|
8
|
5
|
0.625
|
0.0128865979381443
|
|
HP
|
HP:0001265
|
Hyporeflexia
|
0.00184867928909346
|
345
|
13
|
6
|
0.461538461538462
|
0.0173913043478261
|
|
HP
|
HP:0000736
|
Short attention span
|
0.0018759052549927
|
390
|
8
|
5
|
0.625
|
0.0128205128205128
|
|
HP
|
HP:0000504
|
Abnormality of vision
|
0.00197332832255279
|
1208
|
11
|
9
|
0.818181818181818
|
0.00745033112582781
|
|
HP
|
HP:0010935
|
Abnormality of the upper urinary tract
|
0.00197332832255279
|
1208
|
11
|
9
|
0.818181818181818
|
0.00745033112582781
|
|
HP
|
HP:0032263
|
Increased blood pressure
|
0.00197346579610597
|
350
|
13
|
6
|
0.461538461538462
|
0.0171428571428571
|
|
HP
|
HP:0002795
|
Abnormal respiratory system physiology
|
0.00211392812972028
|
1219
|
11
|
9
|
0.818181818181818
|
0.007383100902379
|
|
HP
|
HP:0002094
|
Dyspnea
|
0.00229015069665042
|
360
|
13
|
6
|
0.461538461538462
|
0.0166666666666667
|
|
HP
|
HP:0001000
|
Abnormality of skin pigmentation
|
0.00230503230582258
|
409
|
8
|
5
|
0.625
|
0.0122249388753056
|
|
HP
|
HP:0001627
|
Abnormal heart morphology
|
0.00247901808299734
|
1244
|
11
|
9
|
0.818181818181818
|
0.00723472668810289
|
|
HP
|
HP:0033354
|
Abnormal urine metabolite level
|
0.00268765290789978
|
423
|
8
|
5
|
0.625
|
0.0118203309692671
|
|
HP
|
HP:0004325
|
Decreased body weight
|
0.00321580838097554
|
1285
|
11
|
9
|
0.818181818181818
|
0.00700389105058366
|
|
HP
|
HP:0002353
|
EEG abnormality
|
0.0036846240675061
|
453
|
8
|
5
|
0.625
|
0.011037527593819
|
|
HP
|
HP:0008972
|
Decreased activity of mitochondrial respiratory chain
|
0.00405877662894808
|
84
|
9
|
3
|
0.333333333333333
|
0.0357142857142857
|
|
HP
|
HP:0011922
|
Abnormal activity of mitochondrial respiratory chain
|
0.00418836548112733
|
85
|
9
|
3
|
0.333333333333333
|
0.0352941176470588
|
|
HP
|
HP:0002079
|
Hypoplasia of the corpus callosum
|
0.00429871757724481
|
469
|
8
|
5
|
0.625
|
0.0106609808102345
|
|
HP
|
HP:0000817
|
Poor eye contact
|
0.00430535678454777
|
86
|
9
|
3
|
0.333333333333333
|
0.0348837209302326
|
|
HP
|
HP:0030178
|
Abnormality of central nervous system electrophysiology
|
0.00448891391370732
|
474
|
8
|
5
|
0.625
|
0.0105485232067511
|
|
HP
|
HP:0000365
|
Hearing impairment
|
0.00470108690962704
|
1349
|
11
|
9
|
0.818181818181818
|
0.00667160859896219
|
|
HP
|
HP:0012337
|
Abnormal homeostasis
|
0.00528806142861996
|
1370
|
11
|
9
|
0.818181818181818
|
0.00656934306569343
|
|
HP
|
HP:0000364
|
Hearing abnormality
|
0.00528806142861996
|
1370
|
11
|
9
|
0.818181818181818
|
0.00656934306569343
|
|
HP
|
HP:0030972
|
Abnormal systemic blood pressure
|
0.00528806142861996
|
423
|
13
|
6
|
0.461538461538462
|
0.0141843971631206
|
|
HP
|
HP:0031704
|
Abnormal ear physiology
|
0.0053987648893201
|
1374
|
11
|
9
|
0.818181818181818
|
0.00655021834061135
|
|
HP
|
HP:0002015
|
Dysphagia
|
0.00556784138680062
|
428
|
13
|
6
|
0.461538461538462
|
0.014018691588785
|
|
HP
|
HP:0001608
|
Abnormality of the voice
|
0.00561971363320601
|
429
|
13
|
6
|
0.461538461538462
|
0.013986013986014
|
|
HP
|
HP:0002059
|
Cerebral atrophy
|
0.00618333444788733
|
624
|
13
|
7
|
0.538461538461538
|
0.0112179487179487
|
|
HP
|
HP:0003271
|
Visceromegaly
|
0.0062863766406052
|
626
|
13
|
7
|
0.538461538461538
|
0.0111821086261981
|
|
HP
|
HP:0001254
|
Lethargy
|
0.00629014489583113
|
189
|
11
|
4
|
0.363636363636364
|
0.0211640211640212
|
|
HP
|
HP:0001903
|
Anemia
|
0.00676535234214929
|
528
|
5
|
4
|
0.8
|
0.00757575757575758
|
|
HP
|
HP:0012379
|
Abnormal enzyme/coenzyme activity
|
0.0068182637703217
|
446
|
13
|
6
|
0.461538461538462
|
0.0134529147982063
|
|
HP
|
HP:0007369
|
Atrophy/Degeneration affecting the cerebrum
|
0.00684414534704081
|
636
|
13
|
7
|
0.538461538461538
|
0.0110062893081761
|
|
HP
|
HP:0002415
|
Leukodystrophy
|
0.00695201833128782
|
103
|
9
|
3
|
0.333333333333333
|
0.029126213592233
|
|
HP
|
HP:0001311
|
Abnormal nervous system electrophysiology
|
0.00816903687844447
|
556
|
5
|
4
|
0.8
|
0.00719424460431655
|
|
HP
|
HP:0001877
|
Abnormal erythrocyte morphology
|
0.00909470992187296
|
572
|
5
|
4
|
0.8
|
0.00699300699300699
|
|
HP
|
HP:0008373
|
Puberty and gonadal disorders
|
0.00956913559552323
|
580
|
5
|
4
|
0.8
|
0.00689655172413793
|
|
HP
|
HP:0001438
|
Abnormal abdomen morphology
|
0.00980621341091518
|
676
|
13
|
7
|
0.538461538461538
|
0.0103550295857988
|
|
HP
|
HP:0031466
|
Impairment in personality functioning
|
0.010708392745499
|
598
|
5
|
4
|
0.8
|
0.00668896321070234
|
|
HP
|
HP:0000735
|
Impaired social interactions
|
0.0109002779868478
|
121
|
9
|
3
|
0.333333333333333
|
0.0247933884297521
|
|
HP
|
HP:0001943
|
Hypoglycemia
|
0.0114696905894328
|
224
|
11
|
4
|
0.363636363636364
|
0.0178571428571429
|
|
HP
|
HP:0012444
|
Brain atrophy
|
0.0114696905894328
|
695
|
13
|
7
|
0.538461538461538
|
0.0100719424460432
|
|
HP
|
HP:0000496
|
Abnormality of eye movement
|
0.0118503886209101
|
1523
|
11
|
9
|
0.818181818181818
|
0.00590938936309915
|
|
HP
|
HP:0030680
|
Abnormality of cardiovascular system morphology
|
0.0121323558608878
|
1528
|
11
|
9
|
0.818181818181818
|
0.00589005235602094
|
|
HP
|
HP:0007370
|
Aplasia/Hypoplasia of the corpus callosum
|
0.0129386602998605
|
710
|
13
|
7
|
0.538461538461538
|
0.00985915492957747
|
|
HP
|
HP:0001315
|
Reduced tendon reflexes
|
0.0133327532186914
|
510
|
13
|
6
|
0.461538461538462
|
0.0117647058823529
|
|
HP
|
HP:0004323
|
Abnormality of body weight
|
0.0137136404980014
|
1553
|
11
|
9
|
0.818181818181818
|
0.00579523502897617
|
|
HP
|
HP:0011458
|
Abdominal symptom
|
0.0138141937732343
|
1555
|
11
|
9
|
0.818181818181818
|
0.00578778135048231
|
|
HP
|
HP:0000618
|
Blindness
|
0.0141552931858882
|
266
|
10
|
4
|
0.4
|
0.0150375939849624
|
|
HP
|
HP:0001270
|
Motor delay
|
0.014341573309841
|
651
|
5
|
4
|
0.8
|
0.00614439324116743
|
|
HP
|
HP:0002977
|
Aplasia/Hypoplasia involving the central nervous system
|
0.014341573309841
|
1564
|
11
|
9
|
0.818181818181818
|
0.00575447570332481
|
|
HP
|
HP:0410042
|
Abnormal liver morphology
|
0.0143695507365609
|
725
|
13
|
7
|
0.538461538461538
|
0.0096551724137931
|
|
HP
|
HP:0011015
|
Abnormal blood glucose concentration
|
0.0159374919435881
|
247
|
11
|
4
|
0.363636363636364
|
0.0161943319838057
|
|
HP
|
HP:0001713
|
Abnormal cardiac ventricle morphology
|
0.0173261002682864
|
539
|
13
|
6
|
0.461538461538462
|
0.0111317254174397
|
|
HP
|
HP:0011805
|
Abnormal skeletal muscle morphology
|
0.0173261002682864
|
1604
|
11
|
9
|
0.818181818181818
|
0.00561097256857855
|
|
HP
|
HP:0007364
|
Aplasia/Hypoplasia of the cerebrum
|
0.0174823054103594
|
1346
|
5
|
5
|
1
|
0.0037147102526003
|
|
HP
|
HP:0011362
|
Abnormal hair quantity
|
0.0199520355353516
|
713
|
5
|
4
|
0.8
|
0.00561009817671809
|
|
HP
|
HP:0000080
|
Abnormality of reproductive system physiology
|
0.0202166113784408
|
716
|
5
|
4
|
0.8
|
0.00558659217877095
|
|
HP
|
HP:0007367
|
Atrophy/Degeneration affecting the central nervous system
|
0.0211116911036283
|
777
|
10
|
6
|
0.6
|
0.00772200772200772
|
|
HP
|
HP:0001273
|
Abnormal corpus callosum morphology
|
0.0224744063428549
|
785
|
13
|
7
|
0.538461538461538
|
0.0089171974522293
|
|
HP
|
HP:0002242
|
Abnormal intestine morphology
|
0.022921245667055
|
742
|
5
|
4
|
0.8
|
0.00539083557951483
|
|
HP
|
HP:0033353
|
Abnormal blood vessel morphology
|
0.0246797814036411
|
799
|
13
|
7
|
0.538461538461538
|
0.00876095118898623
|
|
HP
|
HP:0011024
|
Abnormality of the gastrointestinal tract
|
0.0246797814036411
|
1504
|
10
|
8
|
0.8
|
0.00531914893617021
|
|
HP
|
HP:0025015
|
Abnormal vascular morphology
|
0.0250781219311987
|
802
|
13
|
7
|
0.538461538461538
|
0.00872817955112219
|
|
HP
|
HP:0001250
|
Seizure
|
0.0258444669139776
|
1694
|
11
|
9
|
0.818181818181818
|
0.00531286894923259
|
|
HP
|
HP:0000079
|
Abnormality of the urinary system
|
0.0261357527407787
|
1697
|
11
|
9
|
0.818181818181818
|
0.00530347672362994
|
|
HP
|
HP:0012189
|
Hodgkin lymphoma
|
0.0269587308973258
|
23
|
130
|
4
|
0.0307692307692308
|
0.173913043478261
|
|
HP
|
HP:0011442
|
Abnormal central motor function
|
0.0274881288962064
|
1709
|
11
|
9
|
0.818181818181818
|
0.00526623756582797
|
|
HP
|
HP:0100025
|
Overfriendliness
|
0.0276096877478838
|
11
|
139
|
3
|
0.0215827338129496
|
0.272727272727273
|
|
HP
|
HP:0012433
|
Abnormal social behavior
|
0.0278857195412316
|
174
|
9
|
3
|
0.333333333333333
|
0.0172413793103448
|
|
HP
|
HP:0002060
|
Abnormal cerebral morphology
|
0.02828415088259
|
1717
|
11
|
9
|
0.818181818181818
|
0.0052417006406523
|
|
HP
|
HP:0002086
|
Abnormality of the respiratory system
|
0.0283337233859049
|
1718
|
11
|
9
|
0.818181818181818
|
0.00523864959254948
|
|
HP
|
HP:0002421
|
Poor head control
|
0.0299579658175284
|
179
|
9
|
3
|
0.333333333333333
|
0.0167597765363128
|
|
HP
|
HP:0100547
|
Abnormality of forebrain morphology
|
0.0320423440553646
|
1746
|
11
|
9
|
0.818181818181818
|
0.00515463917525773
|
|
HP
|
HP:0001263
|
Global developmental delay
|
0.0368017649896115
|
1601
|
10
|
8
|
0.8
|
0.00499687695190506
|
|
HP
|
HP:0001510
|
Growth delay
|
0.0375183783329304
|
1606
|
10
|
8
|
0.8
|
0.0049813200498132
|
|
HP
|
HP:0031703
|
Abnormal ear morphology
|
0.0409372784644541
|
1806
|
11
|
9
|
0.818181818181818
|
0.00498338870431894
|
|
HP
|
HP:0011298
|
Prominent digit pad
|
0.0410726626325295
|
38
|
141
|
5
|
0.0354609929078014
|
0.131578947368421
|
|
HP
|
HP:0001212
|
Prominent fingertip pads
|
0.0410726626325295
|
38
|
141
|
5
|
0.0354609929078014
|
0.131578947368421
|
|
HP
|
HP:0000708
|
Behavioral abnormality
|
0.0411558914671521
|
1631
|
10
|
8
|
0.8
|
0.00490496627835684
|
|
HP
|
HP:0002352
|
Leukoencephalopathy
|
0.0423257501268324
|
205
|
9
|
3
|
0.333333333333333
|
0.0146341463414634
|
|
HP
|
HP:0025270
|
Abnormality of esophagus physiology
|
0.0432120686765044
|
657
|
13
|
6
|
0.461538461538462
|
0.0091324200913242
|
|
HP
|
HP:0100022
|
Abnormality of movement
|
0.0444821447698154
|
1829
|
11
|
9
|
0.818181818181818
|
0.00492072170585019
|
|
HP
|
HP:0001211
|
Abnormal fingertip morphology
|
0.0453346272797557
|
72
|
141
|
7
|
0.049645390070922
|
0.0972222222222222
|
|
HP
|
HP:0002500
|
Abnormality of the cerebral white matter
|
0.0457443867968517
|
910
|
5
|
4
|
0.8
|
0.0043956043956044
|
|
HP
|
HP:0012649
|
Increased inflammatory response
|
0.0462622379300954
|
914
|
5
|
4
|
0.8
|
0.00437636761487965
|
|
HP
|
HP:0012647
|
Abnormal inflammatory response
|
0.0462622379300954
|
914
|
5
|
4
|
0.8
|
0.00437636761487965
|
|
HP
|
HP:0001347
|
Hyperreflexia
|
0.0485560346650084
|
675
|
13
|
6
|
0.461538461538462
|
0.00888888888888889
|
|
HP
|
HP:0002363
|
Abnormal brainstem morphology
|
0.0496727999974027
|
219
|
9
|
3
|
0.333333333333333
|
0.0136986301369863
|
|
HPA
|
HPA:0090193
|
cerebellum; cells in molecular layer[High]
|
0.000119203992220787
|
874
|
130
|
29
|
0.223076923076923
|
0.0331807780320366
|
|
HPA
|
HPA:0301172
|
lung; alveolar cells[≥Medium]
|
0.000119203992220787
|
2863
|
114
|
55
|
0.482456140350877
|
0.0192106182326231
|
|
HPA
|
HPA:0160253
|
endometrium 1; cells in endometrial stroma[High]
|
0.000367817994464946
|
393
|
138
|
18
|
0.130434782608696
|
0.0458015267175573
|
|
HPA
|
HPA:0030073
|
appendix; lymphoid tissue[High]
|
0.000413458342832867
|
803
|
140
|
27
|
0.192857142857143
|
0.0336239103362391
|
|
HPA
|
HPA:0490693
|
smooth muscle; smooth muscle cells[High]
|
0.000574047572776009
|
589
|
130
|
21
|
0.161538461538462
|
0.0356536502546689
|
|
HPA
|
HPA:0330223
|
oral mucosa; squamous epithelial cells[High]
|
0.000574047572776009
|
1112
|
67
|
20
|
0.298507462686567
|
0.0179856115107914
|
|
HPA
|
HPA:0100123
|
cerebral cortex; glial cells[High]
|
0.00109563050663942
|
683
|
130
|
22
|
0.169230769230769
|
0.0322108345534407
|
|
HPA
|
HPA:0460643
|
skin 1; Langerhans[High]
|
0.00109563050663942
|
734
|
138
|
24
|
0.173913043478261
|
0.0326975476839237
|
|
HPA
|
HPA:0600433
|
tonsil; germinal center cells[High]
|
0.0015805377463549
|
1179
|
140
|
32
|
0.228571428571429
|
0.0271416454622561
|
|
HPA
|
HPA:0090192
|
cerebellum; cells in molecular layer[≥Medium]
|
0.0015805377463549
|
2699
|
113
|
48
|
0.424778761061947
|
0.0177843645794739
|
|
HPA
|
HPA:0600442
|
tonsil; non-germinal center cells[≥Medium]
|
0.00167605823485558
|
4079
|
114
|
64
|
0.56140350877193
|
0.0156901201274822
|
|
HPA
|
HPA:0110223
|
cervix, uterine; squamous epithelial cells[High]
|
0.00167605823485558
|
986
|
67
|
17
|
0.253731343283582
|
0.0172413793103448
|
|
HPA
|
HPA:0050011
|
breast; adipocytes[≥Low]
|
0.00167605823485558
|
2725
|
130
|
53
|
0.407692307692308
|
0.0194495412844037
|
|
HPA
|
HPA:0460673
|
skin 1; melanocytes[High]
|
0.00167605823485558
|
848
|
138
|
25
|
0.181159420289855
|
0.0294811320754717
|
|
HPA
|
HPA:0050012
|
breast; adipocytes[≥Medium]
|
0.00167605823485558
|
1445
|
124
|
33
|
0.266129032258065
|
0.0228373702422145
|
|
HPA
|
HPA:0340463
|
ovary; ovarian stroma cells[High]
|
0.00167605823485558
|
349
|
130
|
14
|
0.107692307692308
|
0.0401146131805158
|
|
HPA
|
HPA:0090182
|
cerebellum; cells in granular layer[≥Medium]
|
0.00167605823485558
|
2854
|
135
|
57
|
0.422222222222222
|
0.0199719691660827
|
|
HPA
|
HPA:0620223
|
vagina; squamous epithelial cells[High]
|
0.00167605823485558
|
1091
|
67
|
18
|
0.26865671641791
|
0.0164986251145738
|
|
HPA
|
HPA:0380513
|
placenta; decidual cells[High]
|
0.00167605823485558
|
1144
|
130
|
29
|
0.223076923076923
|
0.0253496503496503
|
|
HPA
|
HPA:0360053
|
parathyroid gland; glandular cells[High]
|
0.00167605823485558
|
1486
|
135
|
36
|
0.266666666666667
|
0.0242261103633917
|
|
HPA
|
HPA:0090162
|
cerebellum; Purkinje cells[≥Medium]
|
0.00167605823485558
|
4047
|
67
|
41
|
0.611940298507463
|
0.0101309612058315
|
|
HPA
|
HPA:0310433
|
lymph node; germinal center cells[High]
|
0.00187289903984787
|
908
|
140
|
26
|
0.185714285714286
|
0.0286343612334802
|
|
HPA
|
HPA:0100121
|
cerebral cortex; glial cells[≥Low]
|
0.00187289903984787
|
4804
|
68
|
46
|
0.676470588235294
|
0.00957535387177352
|
|
HPA
|
HPA:0100133
|
cerebral cortex; neuronal cells[High]
|
0.00187289903984787
|
1785
|
67
|
24
|
0.358208955223881
|
0.0134453781512605
|
|
HPA
|
HPA:0170252
|
endometrium 2; cells in endometrial stroma[≥Medium]
|
0.00223834372927895
|
1722
|
87
|
28
|
0.32183908045977
|
0.016260162601626
|
|
HPA
|
HPA:0110053
|
cervix, uterine; glandular cells[High]
|
0.00225524041223775
|
1135
|
67
|
18
|
0.26865671641791
|
0.0158590308370044
|
|
HPA
|
HPA:0170251
|
endometrium 2; cells in endometrial stroma[≥Low]
|
0.00235135279913174
|
3410
|
114
|
55
|
0.482456140350877
|
0.0161290322580645
|
|
HPA
|
HPA:0170253
|
endometrium 2; cells in endometrial stroma[High]
|
0.00235135279913174
|
326
|
130
|
13
|
0.1
|
0.0398773006134969
|
|
HPA
|
HPA:0090191
|
cerebellum; cells in molecular layer[≥Low]
|
0.002417282208109
|
4336
|
103
|
60
|
0.58252427184466
|
0.0138376383763838
|
|
HPA
|
HPA:0600443
|
tonsil; non-germinal center cells[High]
|
0.00252048791357049
|
1271
|
67
|
19
|
0.283582089552239
|
0.014948859166011
|
|
HPA
|
HPA:0310443
|
lymph node; non-germinal center cells[High]
|
0.00252048791357049
|
1169
|
140
|
30
|
0.214285714285714
|
0.0256629597946963
|
|
HPA
|
HPA:0460663
|
skin 1; keratinocytes[High]
|
0.00252048791357049
|
848
|
138
|
24
|
0.173913043478261
|
0.0283018867924528
|
|
HPA
|
HPA:0460642
|
skin 1; Langerhans[≥Medium]
|
0.0027049348257837
|
3123
|
130
|
57
|
0.438461538461538
|
0.0182516810758886
|
|
HPA
|
HPA:0310442
|
lymph node; non-germinal center cells[≥Medium]
|
0.0027049348257837
|
3875
|
135
|
69
|
0.511111111111111
|
0.0178064516129032
|
|
HPA
|
HPA:0460651
|
skin 1; fibroblasts[≥Low]
|
0.0030166916381275
|
4726
|
114
|
69
|
0.605263157894737
|
0.0146000846381718
|
|
HPA
|
HPA:0160252
|
endometrium 1; cells in endometrial stroma[≥Medium]
|
0.00307731640442484
|
2142
|
96
|
34
|
0.354166666666667
|
0.0158730158730159
|
|
HPA
|
HPA:0460653
|
skin 1; fibroblasts[High]
|
0.00307731640442484
|
816
|
104
|
19
|
0.182692307692308
|
0.0232843137254902
|
|
HPA
|
HPA:0530723
|
spleen; cells in white pulp[High]
|
0.00334031805749722
|
811
|
140
|
23
|
0.164285714285714
|
0.0283600493218249
|
|
HPA
|
HPA:0301173
|
lung; alveolar cells[High]
|
0.00423443469043455
|
778
|
130
|
21
|
0.161538461538462
|
0.0269922879177378
|
|
HPA
|
HPA:0610833
|
urinary bladder; urothelial cells[High]
|
0.00423443469043455
|
1969
|
70
|
25
|
0.357142857142857
|
0.0126968004062976
|
|
HPA
|
HPA:0530721
|
spleen; cells in white pulp[≥Low]
|
0.00423443469043455
|
4249
|
133
|
72
|
0.541353383458647
|
0.0169451635678983
|
|
HPA
|
HPA:0100132
|
cerebral cortex; neuronal cells[≥Medium]
|
0.00426158009516745
|
4883
|
68
|
45
|
0.661764705882353
|
0.00921564611918902
|
|
HPA
|
HPA:0530722
|
spleen; cells in white pulp[≥Medium]
|
0.00426158009516745
|
2645
|
63
|
28
|
0.444444444444444
|
0.010586011342155
|
|
HPA
|
HPA:0250122
|
hippocampus; glial cells[≥Medium]
|
0.0043066231698413
|
1926
|
96
|
31
|
0.322916666666667
|
0.0160955347871236
|
|
HPA
|
HPA:0270353
|
kidney; cells in glomeruli[High]
|
0.00431584124473198
|
742
|
138
|
21
|
0.152173913043478
|
0.0283018867924528
|
|
HPA
|
HPA:0360052
|
parathyroid gland; glandular cells[≥Medium]
|
0.00431584124473198
|
4306
|
132
|
72
|
0.545454545454545
|
0.0167208546214584
|
|
HPA
|
HPA:0100122
|
cerebral cortex; glial cells[≥Medium]
|
0.00431584124473198
|
2588
|
96
|
38
|
0.395833333333333
|
0.0146831530139104
|
|
HPA
|
HPA:0340462
|
ovary; ovarian stroma cells[≥Medium]
|
0.00431584124473198
|
2002
|
142
|
43
|
0.302816901408451
|
0.0214785214785215
|
|
HPA
|
HPA:0160053
|
endometrium 1; glandular cells[High]
|
0.00431584124473198
|
1413
|
140
|
33
|
0.235714285714286
|
0.0233545647558386
|
|
HPA
|
HPA:0570773
|
testis; peritubular cells[High]
|
0.00438324862436991
|
192
|
130
|
9
|
0.0692307692307692
|
0.046875
|
|
HPA
|
HPA:0080121
|
caudate; glial cells[≥Low]
|
0.00458216462268177
|
4464
|
68
|
42
|
0.617647058823529
|
0.00940860215053763
|
|
HPA
|
HPA:0570781
|
testis; preleptotene spermatocytes[≥Low]
|
0.00458216462268177
|
1636
|
71
|
22
|
0.309859154929577
|
0.0134474327628362
|
|
HPA
|
HPA:0090161
|
cerebellum; Purkinje cells[≥Low]
|
0.00458216462268177
|
5460
|
71
|
50
|
0.704225352112676
|
0.00915750915750916
|
|
HPA
|
HPA:0390052
|
prostate; glandular cells[≥Medium]
|
0.00458216462268177
|
4699
|
67
|
43
|
0.641791044776119
|
0.0091508831666312
|
|
HPA
|
HPA:0620222
|
vagina; squamous epithelial cells[≥Medium]
|
0.00516731700773594
|
3945
|
114
|
59
|
0.517543859649123
|
0.0149556400506971
|
|
HPA
|
HPA:0080122
|
caudate; glial cells[≥Medium]
|
0.00527337354420143
|
2230
|
142
|
46
|
0.323943661971831
|
0.020627802690583
|
|
HPA
|
HPA:0600223
|
tonsil; squamous epithelial cells[High]
|
0.00527337354420143
|
1442
|
140
|
33
|
0.235714285714286
|
0.0228848821081831
|
|
HPA
|
HPA:0351183
|
pancreas; pancreatic endocrine cells[High]
|
0.00532913280197733
|
758
|
130
|
20
|
0.153846153846154
|
0.0263852242744063
|
|
HPA
|
HPA:0250132
|
hippocampus; neuronal cells[≥Medium]
|
0.00533571281889994
|
4126
|
67
|
39
|
0.582089552238806
|
0.00945225399903054
|
|
HPA
|
HPA:0030072
|
appendix; lymphoid tissue[≥Medium]
|
0.00539438963137677
|
3131
|
56
|
28
|
0.5
|
0.00894282976684765
|
|
HPA
|
HPA:0600432
|
tonsil; germinal center cells[≥Medium]
|
0.00558913199647687
|
3988
|
67
|
38
|
0.567164179104478
|
0.00952858575727182
|
|
HPA
|
HPA:0100203
|
cerebral cortex; endothelial cells[High]
|
0.00572766078353946
|
391
|
130
|
13
|
0.1
|
0.0332480818414322
|
|
HPA
|
HPA:0180053
|
epididymis; glandular cells[High]
|
0.00572766078353946
|
1779
|
130
|
36
|
0.276923076923077
|
0.0202360876897133
|
|
HPA
|
HPA:0600222
|
tonsil; squamous epithelial cells[≥Medium]
|
0.00572766078353946
|
4851
|
115
|
69
|
0.6
|
0.0142238713667285
|
|
HPA
|
HPA:0040083
|
bone marrow; hematopoietic cells[High]
|
0.00572766078353946
|
1376
|
142
|
32
|
0.225352112676056
|
0.0232558139534884
|
|
HPA
|
HPA:0380523
|
placenta; trophoblastic cells[High]
|
0.00576040612899082
|
1998
|
142
|
42
|
0.295774647887324
|
0.021021021021021
|
|
HPA
|
HPA:0241102
|
heart muscle; cardiomyocytes[≥Medium]
|
0.00576040612899082
|
4249
|
70
|
41
|
0.585714285714286
|
0.0096493292539421
|
|
HPA
|
HPA:0170053
|
endometrium 2; glandular cells[High]
|
0.00576040612899082
|
1282
|
140
|
30
|
0.214285714285714
|
0.0234009360374415
|
|
HPA
|
HPA:0190223
|
esophagus; squamous epithelial cells[High]
|
0.00629939577174614
|
1596
|
140
|
35
|
0.25
|
0.0219298245614035
|
|
HPA
|
HPA:0530712
|
spleen; cells in red pulp[≥Medium]
|
0.00642106898752332
|
3049
|
114
|
48
|
0.421052631578947
|
0.015742866513611
|
|
HPA
|
HPA:0490691
|
smooth muscle; smooth muscle cells[≥Low]
|
0.00661311339317372
|
5324
|
67
|
46
|
0.686567164179104
|
0.00864012021036814
|
|
HPA
|
HPA:0490000
|
smooth muscle
|
0.00661311339317372
|
5324
|
67
|
46
|
0.686567164179104
|
0.00864012021036814
|
|
HPA
|
HPA:0570771
|
testis; peritubular cells[≥Low]
|
0.00671120581647114
|
616
|
130
|
17
|
0.130769230769231
|
0.0275974025974026
|
|
HPA
|
HPA:0490692
|
smooth muscle; smooth muscle cells[≥Medium]
|
0.00698675068199096
|
3038
|
67
|
31
|
0.462686567164179
|
0.0102040816326531
|
|
HPA
|
HPA:0110222
|
cervix, uterine; squamous epithelial cells[≥Medium]
|
0.00698675068199096
|
3617
|
67
|
35
|
0.522388059701492
|
0.00967652750898535
|
|
HPA
|
HPA:0460641
|
skin 1; Langerhans[≥Low]
|
0.00736155440943844
|
4634
|
113
|
65
|
0.575221238938053
|
0.0140267587397497
|
|
HPA
|
HPA:0540053
|
stomach 1; glandular cells[High]
|
0.00765744543350306
|
2365
|
142
|
47
|
0.330985915492958
|
0.0198731501057082
|
|
HPA
|
HPA:0340453
|
ovary; follicle cells[High]
|
0.00797635575753874
|
579
|
117
|
15
|
0.128205128205128
|
0.0259067357512953
|
|
HPA
|
HPA:0160251
|
endometrium 1; cells in endometrial stroma[≥Low]
|
0.00849928206970686
|
3962
|
67
|
37
|
0.552238805970149
|
0.00933871781928319
|
|
HPA
|
HPA:0291162
|
liver; cholangiocytes[≥Medium]
|
0.00858387178986558
|
1910
|
108
|
32
|
0.296296296296296
|
0.0167539267015707
|
|
HPA
|
HPA:0570782
|
testis; preleptotene spermatocytes[≥Medium]
|
0.00899137371448999
|
1280
|
67
|
17
|
0.253731343283582
|
0.01328125
|
|
HPA
|
HPA:0590053
|
thyroid gland; glandular cells[High]
|
0.00924005196599657
|
1804
|
70
|
22
|
0.314285714285714
|
0.0121951219512195
|
|
HPA
|
HPA:0440343
|
skeletal muscle; myocytes[High]
|
0.0100991919794901
|
830
|
138
|
21
|
0.152173913043478
|
0.0253012048192771
|
|
HPA
|
HPA:0301171
|
lung; alveolar cells[≥Low]
|
0.0102199677929729
|
5028
|
114
|
69
|
0.605263157894737
|
0.0137231503579952
|
|
HPA
|
HPA:0090183
|
cerebellum; cells in granular layer[High]
|
0.0104833166573709
|
709
|
130
|
18
|
0.138461538461538
|
0.0253878702397743
|
|
HPA
|
HPA:0510701
|
soft tissue 2; peripheral nerve[≥Low]
|
0.0105351798221683
|
3100
|
104
|
44
|
0.423076923076923
|
0.0141935483870968
|
|
HPA
|
HPA:0130202
|
colon; endothelial cells[≥Medium]
|
0.0109309022439869
|
3655
|
132
|
61
|
0.462121212121212
|
0.0166894664842681
|
|
HPA
|
HPA:0050013
|
breast; adipocytes[High]
|
0.0109612578330173
|
284
|
130
|
10
|
0.0769230769230769
|
0.0352112676056338
|
|
HPA
|
HPA:0570813
|
testis; spermatogonia cells[High]
|
0.0112445325095545
|
1281
|
142
|
29
|
0.204225352112676
|
0.0226385636221702
|
|
HPA
|
HPA:0570802
|
testis; sertoli cells[≥Medium]
|
0.0114465533404178
|
1662
|
5
|
4
|
0.8
|
0.00240673886883273
|
|
HPA
|
HPA:0340461
|
ovary; ovarian stroma cells[≥Low]
|
0.0114465533404178
|
3894
|
67
|
36
|
0.537313432835821
|
0.00924499229583975
|
|
HPA
|
HPA:0570772
|
testis; peritubular cells[≥Medium]
|
0.0114612070934495
|
440
|
130
|
13
|
0.1
|
0.0295454545454545
|
|
HPA
|
HPA:0530713
|
spleen; cells in red pulp[High]
|
0.0114612070934495
|
778
|
140
|
20
|
0.142857142857143
|
0.025706940874036
|
|
HPA
|
HPA:0420053
|
salivary gland; glandular cells[High]
|
0.012238224828756
|
1418
|
130
|
29
|
0.223076923076923
|
0.0204513399153738
|
|
HPA
|
HPA:0570811
|
testis; spermatogonia cells[≥Low]
|
0.0133858098174913
|
2732
|
71
|
29
|
0.408450704225352
|
0.0106149341142021
|
|
HPA
|
HPA:0110052
|
cervix, uterine; glandular cells[≥Medium]
|
0.0133858098174913
|
3938
|
67
|
36
|
0.537313432835821
|
0.00914169629253428
|
|
HPA
|
HPA:0190222
|
esophagus; squamous epithelial cells[≥Medium]
|
0.0135355479738891
|
4937
|
123
|
72
|
0.585365853658537
|
0.0145837553169941
|
|
HPA
|
HPA:0270363
|
kidney; cells in tubules[High]
|
0.0135537006865744
|
2461
|
99
|
35
|
0.353535353535354
|
0.0142218610321008
|
|
HPA
|
HPA:0460652
|
skin 1; fibroblasts[≥Medium]
|
0.0137421527261596
|
3304
|
130
|
55
|
0.423076923076923
|
0.0166464891041162
|
|
HPA
|
HPA:0570801
|
testis; sertoli cells[≥Low]
|
0.0148608606719114
|
2256
|
67
|
24
|
0.358208955223881
|
0.0106382978723404
|
|
HPA
|
HPA:0080123
|
caudate; glial cells[High]
|
0.0151702860135097
|
628
|
130
|
16
|
0.123076923076923
|
0.0254777070063694
|
|
HPA
|
HPA:0350472
|
pancreas; exocrine glandular cells[≥Medium]
|
0.0158899393828485
|
5157
|
68
|
44
|
0.647058823529412
|
0.00853209230172581
|
|
HPA
|
HPA:0010012
|
adipose tissue; adipocytes[≥Medium]
|
0.0159840709779617
|
2168
|
70
|
24
|
0.342857142857143
|
0.011070110701107
|
|
HPA
|
HPA:0510702
|
soft tissue 2; peripheral nerve[≥Medium]
|
0.0159840709779617
|
1477
|
63
|
17
|
0.26984126984127
|
0.011509817197021
|
|
HPA
|
HPA:0241103
|
heart muscle; cardiomyocytes[High]
|
0.016242445489942
|
1113
|
4
|
3
|
0.75
|
0.00269541778975741
|
|
HPA
|
HPA:0080133
|
caudate; neuronal cells[High]
|
0.0164579593967116
|
1059
|
59
|
13
|
0.220338983050847
|
0.012275731822474
|
|
HPA
|
HPA:0430052
|
seminal vesicle; glandular cells[≥Medium]
|
0.0164579593967116
|
5096
|
130
|
77
|
0.592307692307692
|
0.0151098901098901
|
|
HPA
|
HPA:0250121
|
hippocampus; glial cells[≥Low]
|
0.0165834991942433
|
4033
|
142
|
69
|
0.485915492957746
|
0.0171088519712373
|
|
HPA
|
HPA:0090163
|
cerebellum; Purkinje cells[High]
|
0.0172570992568171
|
1527
|
98
|
24
|
0.244897959183673
|
0.0157170923379175
|
|
HPA
|
HPA:0500652
|
soft tissue 1; fibroblasts[≥Medium]
|
0.0178479680573078
|
2649
|
59
|
24
|
0.406779661016949
|
0.00906002265005663
|
|
HPA
|
HPA:0270352
|
kidney; cells in glomeruli[≥Medium]
|
0.0178585398495659
|
2914
|
114
|
44
|
0.385964912280702
|
0.0150995195607412
|
|
HPA
|
HPA:0250133
|
hippocampus; neuronal cells[High]
|
0.0184435957366075
|
1210
|
59
|
14
|
0.23728813559322
|
0.0115702479338843
|
|
HPA
|
HPA:0080131
|
caudate; neuronal cells[≥Low]
|
0.0187339685728989
|
6042
|
68
|
49
|
0.720588235294118
|
0.00810989738497186
|
|
HPA
|
HPA:0530711
|
spleen; cells in red pulp[≥Low]
|
0.0187702121423994
|
5195
|
99
|
61
|
0.616161616161616
|
0.0117420596727623
|
|
HPA
|
HPA:0090181
|
cerebellum; cells in granular layer[≥Low]
|
0.0188349807670928
|
4438
|
70
|
40
|
0.571428571428571
|
0.00901306894997747
|
|
HPA
|
HPA:0010013
|
adipose tissue; adipocytes[High]
|
0.0198045729977897
|
427
|
130
|
12
|
0.0923076923076923
|
0.0281030444964871
|
|
HPA
|
HPA:0570763
|
testis; pachytene spermatocytes[High]
|
0.0198045729977897
|
822
|
142
|
20
|
0.140845070422535
|
0.024330900243309
|
|
HPA
|
HPA:0130203
|
colon; endothelial cells[High]
|
0.0198045729977897
|
776
|
130
|
18
|
0.138461538461538
|
0.0231958762886598
|
|
HPA
|
HPA:0570783
|
testis; preleptotene spermatocytes[High]
|
0.0204612881104576
|
603
|
142
|
16
|
0.112676056338028
|
0.0265339966832504
|
|
HPA
|
HPA:0570761
|
testis; pachytene spermatocytes[≥Low]
|
0.0204612881104576
|
2077
|
87
|
27
|
0.310344827586207
|
0.0129995185363505
|
|
HPA
|
HPA:0500703
|
soft tissue 1; peripheral nerve[High]
|
0.0204612881104576
|
225
|
130
|
8
|
0.0615384615384615
|
0.0355555555555556
|
|
HPA
|
HPA:0351182
|
pancreas; pancreatic endocrine cells[≥Medium]
|
0.0204790724649039
|
3523
|
115
|
51
|
0.443478260869565
|
0.0144762986091399
|
|
HPA
|
HPA:0350473
|
pancreas; exocrine glandular cells[High]
|
0.0214734069711479
|
1879
|
142
|
37
|
0.26056338028169
|
0.0196913251729643
|
|
HPA
|
HPA:0460662
|
skin 1; keratinocytes[≥Medium]
|
0.0214734069711479
|
3893
|
122
|
58
|
0.475409836065574
|
0.0148985358335474
|
|
HPA
|
HPA:0360051
|
parathyroid gland; glandular cells[≥Low]
|
0.0218278580313833
|
5767
|
68
|
47
|
0.691176470588235
|
0.00814981792959944
|
|
HPA
|
HPA:0360000
|
parathyroid gland
|
0.0218278580313833
|
5767
|
68
|
47
|
0.691176470588235
|
0.00814981792959944
|
|
HPA
|
HPA:0320103
|
nasopharynx; respiratory epithelial cells[High]
|
0.0231140437039426
|
1844
|
117
|
31
|
0.264957264957265
|
0.0168112798264642
|
|
HPA
|
HPA:0330222
|
oral mucosa; squamous epithelial cells[≥Medium]
|
0.0237717288803963
|
4254
|
122
|
62
|
0.508196721311475
|
0.0145745181006112
|
|
HPA
|
HPA:0500651
|
soft tissue 1; fibroblasts[≥Low]
|
0.0272016331404723
|
4156
|
67
|
36
|
0.537313432835821
|
0.00866217516843118
|
|
HPA
|
HPA:0340452
|
ovary; follicle cells[≥Medium]
|
0.0272016331404723
|
2183
|
117
|
35
|
0.299145299145299
|
0.016032982134677
|
|
HPA
|
HPA:0310432
|
lymph node; germinal center cells[≥Medium]
|
0.0279480219273566
|
3209
|
142
|
56
|
0.394366197183099
|
0.0174509192894983
|
|
HPA
|
HPA:0340451
|
ovary; follicle cells[≥Low]
|
0.027981341174063
|
3228
|
124
|
50
|
0.403225806451613
|
0.0154894671623296
|
|
HPA
|
HPA:0510652
|
soft tissue 2; fibroblasts[≥Medium]
|
0.027981341174063
|
2543
|
130
|
43
|
0.330769230769231
|
0.0169091624066064
|
|
HPA
|
HPA:0570812
|
testis; spermatogonia cells[≥Medium]
|
0.0281399631602137
|
2285
|
67
|
23
|
0.343283582089552
|
0.0100656455142232
|
|
HPA
|
HPA:0460671
|
skin 1; melanocytes[≥Low]
|
0.028235164361195
|
5378
|
114
|
70
|
0.614035087719298
|
0.013015991074749
|
|
HPA
|
HPA:0060103
|
bronchus; respiratory epithelial cells[High]
|
0.0284876050318584
|
1890
|
67
|
20
|
0.298507462686567
|
0.0105820105820106
|
|
HPA
|
HPA:0470683
|
skin 2; epidermal cells[High]
|
0.0284876050318584
|
1346
|
117
|
24
|
0.205128205128205
|
0.0178306092124814
|
|
HPA
|
HPA:0550053
|
stomach 2; glandular cells[High]
|
0.0284876050318584
|
2350
|
5
|
4
|
0.8
|
0.00170212765957447
|
|
HPA
|
HPA:0480053
|
small intestine; glandular cells[High]
|
0.0293055989130318
|
2723
|
70
|
27
|
0.385714285714286
|
0.00991553433712817
|
|
HPA
|
HPA:0540052
|
stomach 1; glandular cells[≥Medium]
|
0.0294514370672936
|
6201
|
68
|
49
|
0.720588235294118
|
0.00790195129817771
|
|
HPA
|
HPA:0110221
|
cervix, uterine; squamous epithelial cells[≥Low]
|
0.0294514370672936
|
5325
|
65
|
42
|
0.646153846153846
|
0.00788732394366197
|
|
HPA
|
HPA:0020053
|
adrenal gland; glandular cells[High]
|
0.0295779233215499
|
2199
|
70
|
23
|
0.328571428571429
|
0.0104592996816735
|
|
HPA
|
HPA:0270351
|
kidney; cells in glomeruli[≥Low]
|
0.0299701753919168
|
5033
|
133
|
76
|
0.571428571428571
|
0.0151003377707133
|
|
HPA
|
HPA:0390053
|
prostate; glandular cells[High]
|
0.0301608633029024
|
1284
|
130
|
25
|
0.192307692307692
|
0.0194704049844237
|
|
HPA
|
HPA:0510651
|
soft tissue 2; fibroblasts[≥Low]
|
0.0317438772700884
|
4037
|
130
|
62
|
0.476923076923077
|
0.0153579390636611
|
|
HPA
|
HPA:0080132
|
caudate; neuronal cells[≥Medium]
|
0.0324518056713484
|
4022
|
70
|
36
|
0.514285714285714
|
0.00895077076081552
|
|
HPA
|
HPA:0440342
|
skeletal muscle; myocytes[≥Medium]
|
0.0326129612438645
|
3677
|
142
|
62
|
0.436619718309859
|
0.0168615719336416
|
|
HPA
|
HPA:0570803
|
testis; sertoli cells[High]
|
0.0339208484719649
|
536
|
129
|
13
|
0.10077519379845
|
0.0242537313432836
|
|
HPA
|
HPA:0460672
|
skin 1; melanocytes[≥Medium]
|
0.0374180119851921
|
3700
|
119
|
53
|
0.445378151260504
|
0.0143243243243243
|
|
HPA
|
HPA:0100212
|
cerebral cortex; neuropil[≥Medium]
|
0.0374180119851921
|
2588
|
5
|
4
|
0.8
|
0.00154559505409583
|
|
HPA
|
HPA:0510653
|
soft tissue 2; fibroblasts[High]
|
0.0374180119851921
|
481
|
130
|
12
|
0.0923076923076923
|
0.0249480249480249
|
|
HPA
|
HPA:0590052
|
thyroid gland; glandular cells[≥Medium]
|
0.0375982971227702
|
5454
|
68
|
44
|
0.647058823529412
|
0.00806747341400807
|
|
HPA
|
HPA:0050092
|
breast; myoepithelial cells[≥Medium]
|
0.0376255989767558
|
3700
|
114
|
51
|
0.447368421052632
|
0.0137837837837838
|
|
HPA
|
HPA:0130201
|
colon; endothelial cells[≥Low]
|
0.0401971577306355
|
5671
|
133
|
83
|
0.62406015037594
|
0.0146358666901781
|
|
HPA
|
HPA:0510000
|
soft tissue 2
|
0.0401971577306355
|
5082
|
67
|
41
|
0.611940298507463
|
0.0080676898858717
|
|
HPA
|
HPA:0530000
|
spleen
|
0.0401971577306355
|
5756
|
133
|
84
|
0.631578947368421
|
0.014593467685893
|
|
HPA
|
HPA:0220053
|
gallbladder; glandular cells[High]
|
0.0401971577306355
|
2772
|
140
|
48
|
0.342857142857143
|
0.0173160173160173
|
|
HPA
|
HPA:0110051
|
cervix, uterine; glandular cells[≥Low]
|
0.0412254635922015
|
5579
|
114
|
71
|
0.62280701754386
|
0.0127262950349525
|
|
HPA
|
HPA:0210053
|
fallopian tube; glandular cells[High]
|
0.0412254635922015
|
1728
|
142
|
33
|
0.232394366197183
|
0.0190972222222222
|
|
HPA
|
HPA:0400053
|
rectum; glandular cells[High]
|
0.0412254635922015
|
2687
|
70
|
26
|
0.371428571428571
|
0.00967621883141049
|
|
HPA
|
HPA:0291163
|
liver; cholangiocytes[High]
|
0.0412254635922015
|
294
|
96
|
7
|
0.0729166666666667
|
0.0238095238095238
|
|
HPA
|
HPA:0600431
|
tonsil; germinal center cells[≥Low]
|
0.0412254635922015
|
5845
|
133
|
85
|
0.639097744360902
|
0.0145423438836612
|
|
HPA
|
HPA:0440341
|
skeletal muscle; myocytes[≥Low]
|
0.0418257548004333
|
5762
|
67
|
45
|
0.671641791044776
|
0.00780978826796251
|
|
HPA
|
HPA:0440000
|
skeletal muscle
|
0.0418257548004333
|
5762
|
67
|
45
|
0.671641791044776
|
0.00780978826796251
|
|
HPA
|
HPA:0250131
|
hippocampus; neuronal cells[≥Low]
|
0.042035598752854
|
6101
|
67
|
47
|
0.701492537313433
|
0.00770365513850188
|
|
HPA
|
HPA:0291161
|
liver; cholangiocytes[≥Low]
|
0.0424310670009223
|
3867
|
108
|
50
|
0.462962962962963
|
0.012929919834497
|
|
HPA
|
HPA:0320102
|
nasopharynx; respiratory epithelial cells[≥Medium]
|
0.0428265766326405
|
5010
|
130
|
73
|
0.561538461538462
|
0.0145708582834331
|
|
HPA
|
HPA:0241101
|
heart muscle; cardiomyocytes[≥Low]
|
0.0428265766326405
|
6188
|
71
|
50
|
0.704225352112676
|
0.00808015513897867
|
|
HPA
|
HPA:0240000
|
heart muscle
|
0.0428265766326405
|
6188
|
71
|
50
|
0.704225352112676
|
0.00808015513897867
|
|
HPA
|
HPA:0150053
|
duodenum; glandular cells[High]
|
0.0428265766326405
|
2767
|
5
|
4
|
0.8
|
0.00144560896277557
|
|
HPA
|
HPA:0050091
|
breast; myoepithelial cells[≥Low]
|
0.0449004972846787
|
5551
|
124
|
76
|
0.612903225806452
|
0.0136912268059809
|
|
HPA
|
HPA:0030071
|
appendix; lymphoid tissue[≥Low]
|
0.0449004972846787
|
4976
|
67
|
40
|
0.597014925373134
|
0.00803858520900321
|
|
HPA
|
HPA:0550052
|
stomach 2; glandular cells[≥Medium]
|
0.0452336385563851
|
6134
|
67
|
47
|
0.701492537313433
|
0.00766221062927943
|
|
HPA
|
HPA:0600441
|
tonsil; non-germinal center cells[≥Low]
|
0.0452336385563851
|
5966
|
67
|
46
|
0.686567164179104
|
0.00771035869929601
|
|
HPA
|
HPA:0500653
|
soft tissue 1; fibroblasts[High]
|
0.0481394577416987
|
509
|
130
|
12
|
0.0923076923076923
|
0.0235756385068762
|
|
HPA
|
HPA:0610832
|
urinary bladder; urothelial cells[≥Medium]
|
0.0481983769025092
|
5819
|
67
|
45
|
0.671641791044776
|
0.00773328750644441
|
|
HPA
|
HPA:0100211
|
cerebral cortex; neuropil[≥Low]
|
0.0483269582242749
|
4942
|
5
|
5
|
1
|
0.00101173613921489
|
|
HPA
|
HPA:0210052
|
fallopian tube; glandular cells[≥Medium]
|
0.049731451092613
|
5023
|
65
|
39
|
0.6
|
0.00776428429225562
|
|
KEGG
|
KEGG:00190
|
Oxidative phosphorylation
|
3.73679925901831e-14
|
133
|
13
|
10
|
0.769230769230769
|
0.075187969924812
|
|
KEGG
|
KEGG:05415
|
Diabetic cardiomyopathy
|
1.41289811983535e-12
|
203
|
13
|
10
|
0.769230769230769
|
0.0492610837438424
|
|
KEGG
|
KEGG:04714
|
Thermogenesis
|
3.64700197401501e-12
|
232
|
13
|
10
|
0.769230769230769
|
0.0431034482758621
|
|
KEGG
|
KEGG:05012
|
Parkinson disease
|
5.5902551017008e-12
|
249
|
13
|
10
|
0.769230769230769
|
0.0401606425702811
|
|
KEGG
|
KEGG:05020
|
Prion disease
|
1.13127086733018e-11
|
273
|
13
|
10
|
0.769230769230769
|
0.0366300366300366
|
|
KEGG
|
KEGG:05016
|
Huntington disease
|
2.97041657960627e-11
|
306
|
13
|
10
|
0.769230769230769
|
0.0326797385620915
|
|
KEGG
|
KEGG:05014
|
Amyotrophic lateral sclerosis
|
1.40986971575582e-10
|
363
|
13
|
10
|
0.769230769230769
|
0.0275482093663912
|
|
KEGG
|
KEGG:05010
|
Alzheimer disease
|
1.4532790618587e-10
|
369
|
13
|
10
|
0.769230769230769
|
0.02710027100271
|
|
KEGG
|
KEGG:05022
|
Pathways of neurodegeneration - multiple diseases
|
1.5973470756361e-09
|
475
|
13
|
10
|
0.769230769230769
|
0.0210526315789474
|
|
KEGG
|
KEGG:04723
|
Retrograde endocannabinoid signaling
|
2.78225922114854e-06
|
148
|
25
|
7
|
0.28
|
0.0472972972972973
|
|
KEGG
|
KEGG:04260
|
Cardiac muscle contraction
|
4.39343518127938e-05
|
87
|
11
|
4
|
0.363636363636364
|
0.0459770114942529
|
|
KEGG
|
KEGG:01100
|
Metabolic pathways
|
7.99206488605368e-05
|
1490
|
13
|
10
|
0.769230769230769
|
0.00671140939597315
|
|
KEGG
|
KEGG:04932
|
Non-alcoholic fatty liver disease
|
0.000233294567862454
|
150
|
4
|
3
|
0.75
|
0.02
|
|
KEGG
|
KEGG:00310
|
Lysine degradation
|
0.0416794688859661
|
63
|
141
|
5
|
0.0354609929078014
|
0.0793650793650794
|
|
KEGG
|
KEGG:03040
|
Spliceosome
|
0.0476866851043165
|
150
|
116
|
7
|
0.0603448275862069
|
0.0466666666666667
|
|
MIRNA
|
MIRNA:hsa-miR-21-5p
|
hsa-miR-21-5p
|
2.91854952367089e-10
|
607
|
136
|
29
|
0.213235294117647
|
0.0477759472817133
|
|
MIRNA
|
MIRNA:hsa-let-7c-5p
|
hsa-let-7c-5p
|
1.36259540505614e-07
|
514
|
37
|
13
|
0.351351351351351
|
0.0252918287937743
|
|
MIRNA
|
MIRNA:hsa-let-7b-5p
|
hsa-let-7b-5p
|
2.61714220405218e-07
|
1212
|
49
|
20
|
0.408163265306122
|
0.0165016501650165
|
|
MIRNA
|
MIRNA:hsa-let-7e-5p
|
hsa-let-7e-5p
|
3.74598831817253e-06
|
608
|
13
|
8
|
0.615384615384615
|
0.0131578947368421
|
|
MIRNA
|
MIRNA:hsa-miR-744-5p
|
hsa-miR-744-5p
|
4.5503008677993e-06
|
435
|
12
|
7
|
0.583333333333333
|
0.0160919540229885
|
|
MIRNA
|
MIRNA:hsa-miR-296-3p
|
hsa-miR-296-3p
|
9.89489052763985e-06
|
255
|
13
|
6
|
0.461538461538462
|
0.0235294117647059
|
|
MIRNA
|
MIRNA:hsa-let-7a-5p
|
hsa-let-7a-5p
|
9.89489052763985e-06
|
637
|
99
|
19
|
0.191919191919192
|
0.0298273155416013
|
|
MIRNA
|
MIRNA:hsa-miR-17-5p
|
hsa-miR-17-5p
|
1.95068456831909e-05
|
1174
|
131
|
30
|
0.229007633587786
|
0.0255536626916525
|
|
MIRNA
|
MIRNA:hsa-miR-19b-3p
|
hsa-miR-19b-3p
|
0.000147843277782488
|
712
|
128
|
21
|
0.1640625
|
0.0294943820224719
|
|
MIRNA
|
MIRNA:hsa-miR-107
|
hsa-miR-107
|
0.000294829387960669
|
299
|
56
|
9
|
0.160714285714286
|
0.0301003344481605
|
|
MIRNA
|
MIRNA:hsa-miR-144-3p
|
hsa-miR-144-3p
|
0.00039203150231341
|
207
|
137
|
11
|
0.0802919708029197
|
0.0531400966183575
|
|
MIRNA
|
MIRNA:hsa-miR-181b-5p
|
hsa-miR-181b-5p
|
0.00039203150231341
|
370
|
141
|
15
|
0.106382978723404
|
0.0405405405405405
|
|
MIRNA
|
MIRNA:hsa-miR-26a-5p
|
hsa-miR-26a-5p
|
0.00039203150231341
|
457
|
132
|
16
|
0.121212121212121
|
0.0350109409190372
|
|
MIRNA
|
MIRNA:hsa-miR-652-3p
|
hsa-miR-652-3p
|
0.00039203150231341
|
162
|
37
|
6
|
0.162162162162162
|
0.037037037037037
|
|
MIRNA
|
MIRNA:hsa-miR-760
|
hsa-miR-760
|
0.00039203150231341
|
194
|
9
|
4
|
0.444444444444444
|
0.0206185567010309
|
|
MIRNA
|
MIRNA:hsa-miR-877-3p
|
hsa-miR-877-3p
|
0.00039203150231341
|
602
|
113
|
17
|
0.150442477876106
|
0.0282392026578073
|
|
MIRNA
|
MIRNA:hsa-miR-320a
|
hsa-miR-320a
|
0.000509245372733049
|
584
|
8
|
5
|
0.625
|
0.00856164383561644
|
|
MIRNA
|
MIRNA:hsa-miR-103a-3p
|
hsa-miR-103a-3p
|
0.000645377058116709
|
452
|
56
|
10
|
0.178571428571429
|
0.0221238938053097
|
|
MIRNA
|
MIRNA:hsa-miR-92a-3p
|
hsa-miR-92a-3p
|
0.000778084011081801
|
1404
|
103
|
25
|
0.242718446601942
|
0.0178062678062678
|
|
MIRNA
|
MIRNA:hsa-miR-18a-5p
|
hsa-miR-18a-5p
|
0.000967946631455308
|
260
|
61
|
8
|
0.131147540983607
|
0.0307692307692308
|
|
MIRNA
|
MIRNA:hsa-miR-32-5p
|
hsa-miR-32-5p
|
0.00110525953469971
|
563
|
134
|
17
|
0.126865671641791
|
0.0301953818827709
|
|
MIRNA
|
MIRNA:hsa-miR-181d-5p
|
hsa-miR-181d-5p
|
0.00137270640478873
|
283
|
141
|
12
|
0.0851063829787234
|
0.0424028268551237
|
|
MIRNA
|
MIRNA:hsa-miR-155-5p
|
hsa-miR-155-5p
|
0.00147816742790146
|
900
|
97
|
18
|
0.185567010309278
|
0.02
|
|
MIRNA
|
MIRNA:hsa-miR-92b-3p
|
hsa-miR-92b-3p
|
0.00148584279722048
|
707
|
134
|
19
|
0.141791044776119
|
0.0268741159830269
|
|
MIRNA
|
MIRNA:hsa-miR-101-3p
|
hsa-miR-101-3p
|
0.00152440831280284
|
391
|
106
|
12
|
0.113207547169811
|
0.030690537084399
|
|
MIRNA
|
MIRNA:hsa-miR-1-3p
|
hsa-miR-1-3p
|
0.00152440831280284
|
915
|
124
|
21
|
0.169354838709677
|
0.0229508196721311
|
|
MIRNA
|
MIRNA:hsa-miR-342-5p
|
hsa-miR-342-5p
|
0.00152440831280284
|
126
|
67
|
6
|
0.0895522388059701
|
0.0476190476190476
|
|
MIRNA
|
MIRNA:hsa-miR-19a-3p
|
hsa-miR-19a-3p
|
0.00203384908041548
|
569
|
128
|
16
|
0.125
|
0.0281195079086116
|
|
MIRNA
|
MIRNA:hsa-miR-218-5p
|
hsa-miR-218-5p
|
0.00207751688502773
|
814
|
132
|
20
|
0.151515151515152
|
0.0245700245700246
|
|
MIRNA
|
MIRNA:hsa-miR-302c-5p
|
hsa-miR-302c-5p
|
0.00207751688502773
|
143
|
94
|
7
|
0.074468085106383
|
0.048951048951049
|
|
MIRNA
|
MIRNA:hsa-miR-302f
|
hsa-miR-302f
|
0.00212834110191017
|
224
|
134
|
10
|
0.0746268656716418
|
0.0446428571428571
|
|
MIRNA
|
MIRNA:hsa-miR-561-3p
|
hsa-miR-561-3p
|
0.00226347769725859
|
140
|
67
|
6
|
0.0895522388059701
|
0.0428571428571429
|
|
MIRNA
|
MIRNA:hsa-miR-320c
|
hsa-miR-320c
|
0.00227436499098052
|
108
|
128
|
7
|
0.0546875
|
0.0648148148148148
|
|
MIRNA
|
MIRNA:hsa-miR-181a-5p
|
hsa-miR-181a-5p
|
0.00315659659608044
|
543
|
141
|
16
|
0.113475177304965
|
0.0294659300184162
|
|
MIRNA
|
MIRNA:hsa-miR-3154
|
hsa-miR-3154
|
0.00406155369489382
|
147
|
104
|
7
|
0.0673076923076923
|
0.0476190476190476
|
|
MIRNA
|
MIRNA:hsa-miR-181c-5p
|
hsa-miR-181c-5p
|
0.00460535768385412
|
286
|
141
|
11
|
0.0780141843971631
|
0.0384615384615385
|
|
MIRNA
|
MIRNA:hsa-miR-543
|
hsa-miR-543
|
0.00495658263456076
|
105
|
66
|
5
|
0.0757575757575758
|
0.0476190476190476
|
|
MIRNA
|
MIRNA:hsa-miR-196a-5p
|
hsa-miR-196a-5p
|
0.00495658263456076
|
302
|
13
|
4
|
0.307692307692308
|
0.0132450331125828
|
|
MIRNA
|
MIRNA:hsa-miR-16-5p
|
hsa-miR-16-5p
|
0.00495658263456076
|
1556
|
140
|
30
|
0.214285714285714
|
0.019280205655527
|
|
MIRNA
|
MIRNA:hsa-miR-203a-3p
|
hsa-miR-203a-3p
|
0.0062568553374236
|
307
|
115
|
10
|
0.0869565217391304
|
0.0325732899022801
|
|
MIRNA
|
MIRNA:hsa-let-7a-2-3p
|
hsa-let-7a-2-3p
|
0.00626682685904602
|
102
|
115
|
6
|
0.0521739130434783
|
0.0588235294117647
|
|
MIRNA
|
MIRNA:hsa-let-7g-3p
|
hsa-let-7g-3p
|
0.00755829007913682
|
106
|
115
|
6
|
0.0521739130434783
|
0.0566037735849057
|
|
MIRNA
|
MIRNA:hsa-miR-221-3p
|
hsa-miR-221-3p
|
0.00887352485337532
|
366
|
142
|
12
|
0.0845070422535211
|
0.0327868852459016
|
|
MIRNA
|
MIRNA:hsa-miR-223-3p
|
hsa-miR-223-3p
|
0.00895951676001997
|
98
|
130
|
6
|
0.0461538461538462
|
0.0612244897959184
|
|
MIRNA
|
MIRNA:hsa-let-7f-2-3p
|
hsa-let-7f-2-3p
|
0.00895951676001997
|
98
|
130
|
6
|
0.0461538461538462
|
0.0612244897959184
|
|
MIRNA
|
MIRNA:hsa-miR-3143
|
hsa-miR-3143
|
0.00903251270788918
|
92
|
89
|
5
|
0.0561797752808989
|
0.0543478260869565
|
|
MIRNA
|
MIRNA:hsa-miR-4761-3p
|
hsa-miR-4761-3p
|
0.00923564144661634
|
43
|
105
|
4
|
0.0380952380952381
|
0.0930232558139535
|
|
MIRNA
|
MIRNA:hsa-miR-27b-3p
|
hsa-miR-27b-3p
|
0.00926985855497393
|
419
|
91
|
10
|
0.10989010989011
|
0.0238663484486874
|
|
MIRNA
|
MIRNA:hsa-miR-432-3p
|
hsa-miR-432-3p
|
0.00964619185400273
|
68
|
124
|
5
|
0.0403225806451613
|
0.0735294117647059
|
|
MIRNA
|
MIRNA:hsa-miR-196b-5p
|
hsa-miR-196b-5p
|
0.00987517846066772
|
150
|
13
|
3
|
0.230769230769231
|
0.02
|
|
MIRNA
|
MIRNA:hsa-miR-186-5p
|
hsa-miR-186-5p
|
0.0104729235405654
|
749
|
13
|
5
|
0.384615384615385
|
0.00667556742323097
|
|
MIRNA
|
MIRNA:hsa-miR-4302
|
hsa-miR-4302
|
0.0104729235405654
|
137
|
138
|
7
|
0.0507246376811594
|
0.0510948905109489
|
|
MIRNA
|
MIRNA:hsa-miR-200a-3p
|
hsa-miR-200a-3p
|
0.0110448635113068
|
150
|
59
|
5
|
0.0847457627118644
|
0.0333333333333333
|
|
MIRNA
|
MIRNA:hsa-miR-320b
|
hsa-miR-320b
|
0.0116979004056153
|
145
|
134
|
7
|
0.0522388059701493
|
0.0482758620689655
|
|
MIRNA
|
MIRNA:hsa-let-7f-5p
|
hsa-let-7f-5p
|
0.0116979004056153
|
395
|
66
|
8
|
0.121212121212121
|
0.020253164556962
|
|
MIRNA
|
MIRNA:hsa-miR-423-5p
|
hsa-miR-423-5p
|
0.0131016932611679
|
342
|
140
|
11
|
0.0785714285714286
|
0.0321637426900585
|
|
MIRNA
|
MIRNA:hsa-miR-4789-5p
|
hsa-miR-4789-5p
|
0.0132189542707255
|
253
|
131
|
9
|
0.0687022900763359
|
0.0355731225296443
|
|
MIRNA
|
MIRNA:hsa-miR-125b-5p
|
hsa-miR-125b-5p
|
0.0146394890931719
|
427
|
80
|
9
|
0.1125
|
0.0210772833723653
|
|
MIRNA
|
MIRNA:hsa-miR-202-3p
|
hsa-miR-202-3p
|
0.0146394890931719
|
204
|
132
|
8
|
0.0606060606060606
|
0.0392156862745098
|
|
MIRNA
|
MIRNA:hsa-miR-130b-3p
|
hsa-miR-130b-3p
|
0.0151629616870521
|
571
|
131
|
14
|
0.106870229007634
|
0.0245183887915937
|
|
MIRNA
|
MIRNA:hsa-miR-219b-3p
|
hsa-miR-219b-3p
|
0.0151629616870521
|
203
|
134
|
8
|
0.0597014925373134
|
0.0394088669950739
|
|
MIRNA
|
MIRNA:hsa-miR-4668-5p
|
hsa-miR-4668-5p
|
0.0151629616870521
|
402
|
123
|
11
|
0.0894308943089431
|
0.027363184079602
|
|
MIRNA
|
MIRNA:hsa-miR-1185-1-3p
|
hsa-miR-1185-1-3p
|
0.0156224475816249
|
116
|
130
|
6
|
0.0461538461538462
|
0.0517241379310345
|
|
MIRNA
|
MIRNA:hsa-miR-126-5p
|
hsa-miR-126-5p
|
0.0161672152430395
|
118
|
129
|
6
|
0.0465116279069767
|
0.0508474576271186
|
|
MIRNA
|
MIRNA:hsa-miR-6505-5p
|
hsa-miR-6505-5p
|
0.0168370576856855
|
78
|
130
|
5
|
0.0384615384615385
|
0.0641025641025641
|
|
MIRNA
|
MIRNA:hsa-miR-106b-5p
|
hsa-miR-106b-5p
|
0.0169925597535852
|
1084
|
133
|
21
|
0.157894736842105
|
0.0193726937269373
|
|
MIRNA
|
MIRNA:hsa-miR-1185-2-3p
|
hsa-miR-1185-2-3p
|
0.0169925597535852
|
119
|
130
|
6
|
0.0461538461538462
|
0.0504201680672269
|
|
MIRNA
|
MIRNA:hsa-miR-510-3p
|
hsa-miR-510-3p
|
0.0169925597535852
|
166
|
62
|
5
|
0.0806451612903226
|
0.0301204819277108
|
|
MIRNA
|
MIRNA:hsa-miR-491-3p
|
hsa-miR-491-3p
|
0.0182842972162112
|
58
|
103
|
4
|
0.0388349514563107
|
0.0689655172413793
|
|
MIRNA
|
MIRNA:hsa-miR-125a-5p
|
hsa-miR-125a-5p
|
0.0184408606995125
|
269
|
23
|
4
|
0.173913043478261
|
0.0148698884758364
|
|
MIRNA
|
MIRNA:hsa-miR-4795-3p
|
hsa-miR-4795-3p
|
0.0200751169118617
|
122
|
88
|
5
|
0.0568181818181818
|
0.040983606557377
|
|
MIRNA
|
MIRNA:hsa-miR-4311
|
hsa-miR-4311
|
0.020076387575697
|
196
|
115
|
7
|
0.0608695652173913
|
0.0357142857142857
|
|
MIRNA
|
MIRNA:hsa-miR-148a-3p
|
hsa-miR-148a-3p
|
0.020076387575697
|
212
|
138
|
8
|
0.0579710144927536
|
0.0377358490566038
|
|
MIRNA
|
MIRNA:hsa-miR-374a-5p
|
hsa-miR-374a-5p
|
0.020076387575697
|
282
|
80
|
7
|
0.0875
|
0.024822695035461
|
|
MIRNA
|
MIRNA:hsa-miR-519d-3p
|
hsa-miR-519d-3p
|
0.0202714147638046
|
900
|
130
|
18
|
0.138461538461538
|
0.02
|
|
MIRNA
|
MIRNA:hsa-miR-3529-5p
|
hsa-miR-3529-5p
|
0.0202714147638046
|
83
|
132
|
5
|
0.0378787878787879
|
0.0602409638554217
|
|
MIRNA
|
MIRNA:hsa-miR-20a-5p
|
hsa-miR-20a-5p
|
0.0206728423992584
|
1064
|
130
|
20
|
0.153846153846154
|
0.018796992481203
|
|
MIRNA
|
MIRNA:hsa-miR-432-5p
|
hsa-miR-432-5p
|
0.0206728423992584
|
92
|
31
|
3
|
0.0967741935483871
|
0.0326086956521739
|
|
MIRNA
|
MIRNA:hsa-miR-27a-3p
|
hsa-miR-27a-3p
|
0.0206728423992584
|
427
|
124
|
11
|
0.0887096774193548
|
0.0257611241217799
|
|
MIRNA
|
MIRNA:hsa-miR-379-5p
|
hsa-miR-379-5p
|
0.0206728423992584
|
85
|
132
|
5
|
0.0378787878787879
|
0.0588235294117647
|
|
MIRNA
|
MIRNA:hsa-miR-200c-3p
|
hsa-miR-200c-3p
|
0.0206728423992584
|
214
|
14
|
3
|
0.214285714285714
|
0.014018691588785
|
|
MIRNA
|
MIRNA:hsa-miR-1276
|
hsa-miR-1276
|
0.0206728423992584
|
159
|
70
|
5
|
0.0714285714285714
|
0.0314465408805031
|
|
MIRNA
|
MIRNA:hsa-miR-7850-5p
|
hsa-miR-7850-5p
|
0.0206728423992584
|
98
|
66
|
4
|
0.0606060606060606
|
0.0408163265306122
|
|
MIRNA
|
MIRNA:hsa-miR-20b-5p
|
hsa-miR-20b-5p
|
0.0207583506322619
|
911
|
130
|
18
|
0.138461538461538
|
0.0197585071350165
|
|
MIRNA
|
MIRNA:hsa-miR-877-5p
|
hsa-miR-877-5p
|
0.0266072864319292
|
233
|
52
|
5
|
0.0961538461538462
|
0.0214592274678112
|
|
MIRNA
|
MIRNA:hsa-miR-101-5p
|
hsa-miR-101-5p
|
0.0266072864319292
|
68
|
47
|
3
|
0.0638297872340425
|
0.0441176470588235
|
|
MIRNA
|
MIRNA:hsa-miR-4697-3p
|
hsa-miR-4697-3p
|
0.0274123178542541
|
55
|
130
|
4
|
0.0307692307692308
|
0.0727272727272727
|
|
MIRNA
|
MIRNA:hsa-miR-1277-5p
|
hsa-miR-1277-5p
|
0.0291963044992748
|
621
|
134
|
14
|
0.104477611940299
|
0.0225442834138486
|
|
MIRNA
|
MIRNA:hsa-miR-4753-3p
|
hsa-miR-4753-3p
|
0.0292460760215675
|
313
|
80
|
7
|
0.0875
|
0.0223642172523962
|
|
MIRNA
|
MIRNA:hsa-miR-4310
|
hsa-miR-4310
|
0.0295826980057459
|
158
|
47
|
4
|
0.0851063829787234
|
0.0253164556962025
|
|
MIRNA
|
MIRNA:hsa-miR-484
|
hsa-miR-484
|
0.0295826980057459
|
890
|
95
|
14
|
0.147368421052632
|
0.0157303370786517
|
|
MIRNA
|
MIRNA:hsa-miR-7157-5p
|
hsa-miR-7157-5p
|
0.0295826980057459
|
158
|
47
|
4
|
0.0851063829787234
|
0.0253164556962025
|
|
MIRNA
|
MIRNA:hsa-miR-3133
|
hsa-miR-3133
|
0.0305211110217407
|
187
|
135
|
7
|
0.0518518518518519
|
0.0374331550802139
|
|
MIRNA
|
MIRNA:hsa-miR-190a-3p
|
hsa-miR-190a-3p
|
0.0384446417602811
|
644
|
134
|
14
|
0.104477611940299
|
0.0217391304347826
|
|
MIRNA
|
MIRNA:hsa-miR-5011-5p
|
hsa-miR-5011-5p
|
0.0384559088187081
|
648
|
119
|
13
|
0.109243697478992
|
0.0200617283950617
|
|
MIRNA
|
MIRNA:hsa-miR-361-5p
|
hsa-miR-361-5p
|
0.0391449999858185
|
160
|
84
|
5
|
0.0595238095238095
|
0.03125
|
|
MIRNA
|
MIRNA:hsa-miR-195-5p
|
hsa-miR-195-5p
|
0.0392764343122322
|
639
|
107
|
12
|
0.11214953271028
|
0.0187793427230047
|
|
MIRNA
|
MIRNA:hsa-miR-141-3p
|
hsa-miR-141-3p
|
0.0392764343122322
|
145
|
136
|
6
|
0.0441176470588235
|
0.0413793103448276
|
|
MIRNA
|
MIRNA:hsa-miR-25-3p
|
hsa-miR-25-3p
|
0.0405629529495218
|
515
|
133
|
12
|
0.0902255639097744
|
0.0233009708737864
|
|
MIRNA
|
MIRNA:hsa-miR-6766-3p
|
hsa-miR-6766-3p
|
0.0405629529495218
|
38
|
103
|
3
|
0.029126213592233
|
0.0789473684210526
|
|
MIRNA
|
MIRNA:hsa-miR-9-3p
|
hsa-miR-9-3p
|
0.0416627955666607
|
102
|
135
|
5
|
0.037037037037037
|
0.0490196078431373
|
|
MIRNA
|
MIRNA:hsa-miR-1301-3p
|
hsa-miR-1301-3p
|
0.04182658037709
|
196
|
103
|
6
|
0.058252427184466
|
0.0306122448979592
|
|
MIRNA
|
MIRNA:hsa-miR-5590-3p
|
hsa-miR-5590-3p
|
0.04182658037709
|
212
|
128
|
7
|
0.0546875
|
0.0330188679245283
|
|
MIRNA
|
MIRNA:hsa-miR-142-3p
|
hsa-miR-142-3p
|
0.0445071453750205
|
386
|
134
|
10
|
0.0746268656716418
|
0.0259067357512953
|
|
MIRNA
|
MIRNA:hsa-miR-30a-5p
|
hsa-miR-30a-5p
|
0.0467647987692613
|
730
|
61
|
9
|
0.147540983606557
|
0.0123287671232877
|
|
MIRNA
|
MIRNA:hsa-miR-424-3p
|
hsa-miR-424-3p
|
0.0467647987692613
|
32
|
132
|
3
|
0.0227272727272727
|
0.09375
|
|
MIRNA
|
MIRNA:hsa-miR-4782-3p
|
hsa-miR-4782-3p
|
0.0468845820017153
|
41
|
103
|
3
|
0.029126213592233
|
0.0731707317073171
|
|
MIRNA
|
MIRNA:hsa-miR-1180-3p
|
hsa-miR-1180-3p
|
0.0470476396649673
|
67
|
131
|
4
|
0.0305343511450382
|
0.0597014925373134
|
|
MIRNA
|
MIRNA:hsa-miR-4519
|
hsa-miR-4519
|
0.0479585130110815
|
91
|
47
|
3
|
0.0638297872340425
|
0.032967032967033
|
|
MIRNA
|
MIRNA:hsa-miR-455-3p
|
hsa-miR-455-3p
|
0.0495394143405339
|
782
|
48
|
8
|
0.166666666666667
|
0.010230179028133
|
|
MIRNA
|
MIRNA:hsa-miR-6834-3p
|
hsa-miR-6834-3p
|
0.0495394143405339
|
41
|
107
|
3
|
0.0280373831775701
|
0.0731707317073171
|
|
MIRNA
|
MIRNA:hsa-miR-15a-5p
|
hsa-miR-15a-5p
|
0.0495394143405339
|
717
|
113
|
13
|
0.115044247787611
|
0.0181311018131102
|
|
MIRNA
|
MIRNA:hsa-miR-15b-5p
|
hsa-miR-15b-5p
|
0.0495394143405339
|
760
|
107
|
13
|
0.121495327102804
|
0.0171052631578947
|
|
MIRNA
|
MIRNA:hsa-miR-142-5p
|
hsa-miR-142-5p
|
0.0495394143405339
|
225
|
128
|
7
|
0.0546875
|
0.0311111111111111
|
|
MIRNA
|
MIRNA:hsa-miR-551b-5p
|
hsa-miR-551b-5p
|
0.0495394143405339
|
144
|
102
|
5
|
0.0490196078431373
|
0.0347222222222222
|
|
MIRNA
|
MIRNA:hsa-let-7f-1-3p
|
hsa-let-7f-1-3p
|
0.0495394143405339
|
114
|
130
|
5
|
0.0384615384615385
|
0.043859649122807
|
|
MIRNA
|
MIRNA:hsa-miR-98-3p
|
hsa-miR-98-3p
|
0.0495394143405339
|
114
|
130
|
5
|
0.0384615384615385
|
0.043859649122807
|
|
MIRNA
|
MIRNA:hsa-miR-7-2-3p
|
hsa-miR-7-2-3p
|
0.0495394143405339
|
167
|
27
|
3
|
0.111111111111111
|
0.0179640718562874
|
|
MIRNA
|
MIRNA:hsa-miR-603
|
hsa-miR-603
|
0.0495394143405339
|
502
|
124
|
11
|
0.0887096774193548
|
0.0219123505976096
|
|
MIRNA
|
MIRNA:hsa-miR-152-3p
|
hsa-miR-152-3p
|
0.0499906757978355
|
157
|
138
|
6
|
0.0434782608695652
|
0.0382165605095541
|
|
MIRNA
|
MIRNA:hsa-miR-4422
|
hsa-miR-4422
|
0.0499906757978355
|
111
|
135
|
5
|
0.037037037037037
|
0.045045045045045
|
|
REAC
|
REAC:R-HSA-163200
|
Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins.
|
4.4369452202306e-15
|
124
|
13
|
10
|
0.769230769230769
|
0.0806451612903226
|
|
REAC
|
REAC:R-HSA-1428517
|
The citric acid (TCA) cycle and respiratory electron transport
|
5.29448280258136e-15
|
174
|
15
|
11
|
0.733333333333333
|
0.0632183908045977
|
|
REAC
|
REAC:R-HSA-611105
|
Respiratory electron transport
|
4.53338292289273e-14
|
100
|
13
|
9
|
0.692307692307692
|
0.09
|
|
REAC
|
REAC:R-HSA-6799198
|
Complex I biogenesis
|
1.63651476880097e-07
|
55
|
23
|
6
|
0.260869565217391
|
0.109090909090909
|
|
REAC
|
REAC:R-HSA-5628897
|
TP53 Regulates Metabolic Genes
|
0.00017068031407032
|
83
|
4
|
3
|
0.75
|
0.036144578313253
|
|
REAC
|
REAC:R-HSA-3700989
|
Transcriptional Regulation by TP53
|
0.000242240336085346
|
360
|
47
|
10
|
0.212765957446809
|
0.0277777777777778
|
|
REAC
|
REAC:R-HSA-6806003
|
Regulation of TP53 Expression and Degradation
|
0.000346493081535869
|
37
|
130
|
6
|
0.0461538461538462
|
0.162162162162162
|
|
REAC
|
REAC:R-HSA-1430728
|
Metabolism
|
0.000578830489657973
|
2070
|
15
|
11
|
0.733333333333333
|
0.00531400966183575
|
|
REAC
|
REAC:R-HSA-6804760
|
Regulation of TP53 Activity through Methylation
|
0.00109472625760638
|
19
|
31
|
3
|
0.0967741935483871
|
0.157894736842105
|
|
REAC
|
REAC:R-HSA-5633007
|
Regulation of TP53 Activity
|
0.0022221080644475
|
160
|
44
|
6
|
0.136363636363636
|
0.0375
|
|
REAC
|
REAC:R-HSA-6804757
|
Regulation of TP53 Degradation
|
0.00303454839425751
|
36
|
130
|
5
|
0.0384615384615385
|
0.138888888888889
|
|
REAC
|
REAC:R-HSA-72163
|
mRNA Splicing - Major Pathway
|
0.0123289462455004
|
178
|
129
|
9
|
0.0697674418604651
|
0.050561797752809
|
|
REAC
|
REAC:R-HSA-3214841
|
PKMTs methylate histone lysines
|
0.0123289462455004
|
46
|
141
|
5
|
0.0354609929078014
|
0.108695652173913
|
|
REAC
|
REAC:R-HSA-72172
|
mRNA Splicing
|
0.0147733583805827
|
186
|
129
|
9
|
0.0697674418604651
|
0.0483870967741935
|
|
REAC
|
REAC:R-HSA-4839726
|
Chromatin organization
|
0.0350982050166004
|
234
|
142
|
10
|
0.0704225352112676
|
0.0427350427350427
|
|
REAC
|
REAC:R-HSA-3247509
|
Chromatin modifying enzymes
|
0.0350982050166004
|
234
|
142
|
10
|
0.0704225352112676
|
0.0427350427350427
|
|
TF
|
TF:M12277
|
Factor: YY2; motif: NKCSGCCATTTTGN
|
6.51241891848696e-10
|
135
|
96
|
13
|
0.135416666666667
|
0.0962962962962963
|
|
TF
|
TF:M08320
|
Factor: GCMa:HOXB13; motif: RTGCGGGTMATAAAN
|
1.3816191030126e-06
|
837
|
10
|
8
|
0.8
|
0.00955794504181601
|
|
TF
|
TF:M08331
|
Factor: GCMb:FOXI1; motif: RTAAATANGGGNN
|
2.70705886959237e-05
|
45
|
13
|
4
|
0.307692307692308
|
0.0888888888888889
|
|
TF
|
TF:M04043
|
Factor: MYBL1; motif: ACCGTTAACSGY
|
2.70705886959237e-05
|
24
|
5
|
3
|
0.6
|
0.125
|
|
TF
|
TF:M08319
|
Factor: GCMa:HOXA13; motif: RTGCGGGTAATAAA
|
3.68457986678388e-05
|
455
|
10
|
6
|
0.6
|
0.0131868131868132
|
|
TF
|
TF:M05660
|
Factor: ZNF286B; motif: NTTGGCGGATGM
|
0.00013191364354754
|
51
|
5
|
3
|
0.6
|
0.0588235294117647
|
|
TF
|
TF:M04312
|
Factor: EMX1; motif: NNTAATTANN
|
0.00013191364354754
|
2772
|
124
|
40
|
0.32258064516129
|
0.0144300144300144
|
|
TF
|
TF:M05958
|
Factor: ZNF286; motif: NTTGGCGGATGM
|
0.00013191364354754
|
51
|
5
|
3
|
0.6
|
0.0588235294117647
|
|
TF
|
TF:M08320_1
|
Factor: GCMa:HOXB13; motif: RTGCGGGTMATAAAN; match class: 1
|
0.00013191364354754
|
22
|
10
|
3
|
0.3
|
0.136363636363636
|
|
TF
|
TF:M07124
|
Factor: AML3; motif: KNKNTYTGTGGTTTK
|
0.000320945186665694
|
1440
|
65
|
18
|
0.276923076923077
|
0.0125
|
|
TF
|
TF:M11042
|
Factor: HMBOX1; motif: NCTAGTTAN
|
0.000320945186665694
|
138
|
25
|
5
|
0.2
|
0.036231884057971
|
|
TF
|
TF:M10679
|
Factor: HPX42B; motif: NNYAATTANN
|
0.000320945186665694
|
2139
|
124
|
33
|
0.266129032258065
|
0.0154277699859748
|
|
TF
|
TF:M08704
|
Factor: GCMa:HOXA2; motif: RTRNGGGYNTAATNN
|
0.000320945186665694
|
342
|
11
|
5
|
0.454545454545455
|
0.0146198830409357
|
|
TF
|
TF:M00095_1
|
Factor: CDP; motif: CCAATAATCGAT; match class: 1
|
0.000320945186665694
|
2983
|
32
|
17
|
0.53125
|
0.00569896077774053
|
|
TF
|
TF:M04320
|
Factor: EVX1; motif: NNTNATTANN
|
0.000320945186665694
|
3270
|
104
|
38
|
0.365384615384615
|
0.0116207951070336
|
|
TF
|
TF:M08473_1
|
Factor: FOXJ2:HOXB13; motif: TTTTATNDGNMAACA; match class: 1
|
0.000373962154732822
|
155
|
44
|
6
|
0.136363636363636
|
0.0387096774193548
|
|
TF
|
TF:M00023_1
|
Factor: HOXA5; motif: TGCNHNCWYCCYCATTAKTGNDCNMNHYCN; match class: 1
|
0.000373962154732822
|
450
|
25
|
7
|
0.28
|
0.0155555555555556
|
|
TF
|
TF:M04327_1
|
Factor: GSX1; motif: NNYMATTANN; match class: 1
|
0.000373962154732822
|
5540
|
124
|
60
|
0.483870967741935
|
0.0108303249097473
|
|
TF
|
TF:M08326
|
Factor: GCMb:Prrxl1; motif: ATRCGGGTNATTAN
|
0.000373962154732822
|
614
|
13
|
6
|
0.461538461538462
|
0.00977198697068404
|
|
TF
|
TF:M01235
|
Factor: ipf1; motif: NVSTAATTAC
|
0.000373962154732822
|
3518
|
74
|
31
|
0.418918918918919
|
0.00881182490051165
|
|
TF
|
TF:M12285_1
|
Factor: ZNF647; motif: NTAGGCCTAN; match class: 1
|
0.000373962154732822
|
429
|
10
|
5
|
0.5
|
0.0116550116550117
|
|
TF
|
TF:M00137_1
|
Factor: Oct-1; motif: NNNRTAATNANNN; match class: 1
|
0.000373962154732822
|
2173
|
13
|
9
|
0.692307692307692
|
0.00414173953060285
|
|
TF
|
TF:M04321
|
Factor: EVX2; motif: NNTNATTANN
|
0.000378255877196254
|
2980
|
107
|
36
|
0.336448598130841
|
0.0120805369127517
|
|
TF
|
TF:M01362
|
Factor: CART1; motif: NGNNYTAATTARTNNNN
|
0.000378255877196254
|
4558
|
116
|
50
|
0.431034482758621
|
0.0109697235629662
|
|
TF
|
TF:M10948_1
|
Factor: rax; motif: NNCRTTAN; match class: 1
|
0.00038333158266863
|
5245
|
79
|
41
|
0.518987341772152
|
0.00781696854146807
|
|
TF
|
TF:M02044_1
|
Factor: YY1; motif: GCCGCCATTTTG; match class: 1
|
0.000406610516636706
|
1198
|
116
|
22
|
0.189655172413793
|
0.0183639398998331
|
|
TF
|
TF:M11594
|
Factor: GCMa; motif: ATGNGGGYR
|
0.000428508486540388
|
651
|
13
|
6
|
0.461538461538462
|
0.00921658986175115
|
|
TF
|
TF:M03924
|
Factor: YY1; motif: NNCGCCATTNN
|
0.000467754573566716
|
10084
|
100
|
74
|
0.74
|
0.00733835779452598
|
|
TF
|
TF:M08020
|
Factor: YY2; motif: NNCCGCCATTW
|
0.000467754573566716
|
1331
|
99
|
21
|
0.212121212121212
|
0.0157776108189331
|
|
TF
|
TF:M10119
|
Factor: REX-1; motif: GGCMGCCATTTT
|
0.000467754573566716
|
1888
|
89
|
24
|
0.269662921348315
|
0.0127118644067797
|
|
TF
|
TF:M01351
|
Factor: hoxa9; motif: NCGGYCATWAAAWTANW
|
0.000467754573566716
|
2159
|
45
|
17
|
0.377777777777778
|
0.0078740157480315
|
|
TF
|
TF:M01013
|
Factor: ipf1; motif: TSNGYCATTANNNNC
|
0.000467754573566716
|
2498
|
10
|
8
|
0.8
|
0.00320256204963971
|
|
TF
|
TF:M12271
|
Factor: ZNF339; motif: NNACCGTTANNNN
|
0.000468064271790328
|
830
|
75
|
14
|
0.186666666666667
|
0.0168674698795181
|
|
TF
|
TF:M10750_1
|
Factor: gsh-1; motif: NTMATNRN; match class: 1
|
0.000468064271790328
|
3954
|
116
|
45
|
0.387931034482759
|
0.0113808801213961
|
|
TF
|
TF:M10663
|
Factor: LBX2; motif: NYYAATTANN
|
0.000477714378267642
|
2267
|
124
|
33
|
0.266129032258065
|
0.0145566828407587
|
|
TF
|
TF:M12310
|
Factor: ZNF597; motif: NGCCGCCATTTTGN
|
0.000549019710727182
|
437
|
141
|
14
|
0.099290780141844
|
0.0320366132723112
|
|
TF
|
TF:M08278
|
Factor: Erm:HOXA2; motif: RNCGGAWGTMATTA
|
0.000554411065108844
|
124
|
17
|
4
|
0.235294117647059
|
0.032258064516129
|
|
TF
|
TF:M04517_1
|
Factor: E2F2; motif: AAAATGGCGCCATTTT; match class: 1
|
0.000554411065108844
|
3501
|
81
|
32
|
0.395061728395062
|
0.00914024564410169
|
|
TF
|
TF:M04327
|
Factor: GSX1; motif: NNYMATTANN
|
0.000584716014598148
|
5709
|
124
|
60
|
0.483870967741935
|
0.0105097214923805
|
|
TF
|
TF:M04349
|
Factor: HOXB5; motif: NNYMATTANN
|
0.000641012552308644
|
5728
|
124
|
60
|
0.483870967741935
|
0.0104748603351955
|
|
TF
|
TF:M03925
|
Factor: YY2; motif: NCCGCCATNTY
|
0.000703941149950518
|
4641
|
115
|
49
|
0.426086956521739
|
0.0105580693815988
|
|
TF
|
TF:M02115_1
|
Factor: Prx2; motif: TYAWAKTAA; match class: 1
|
0.000820024852491879
|
684
|
9
|
5
|
0.555555555555556
|
0.00730994152046784
|
|
TF
|
TF:M00426_1
|
Factor: E2F; motif: TTTSGCGS; match class: 1
|
0.000863955647976115
|
5830
|
113
|
56
|
0.495575221238938
|
0.00960548885077187
|
|
TF
|
TF:M02115
|
Factor: Prx2; motif: TYAWAKTAA
|
0.000894947159448298
|
3528
|
29
|
16
|
0.551724137931034
|
0.00453514739229025
|
|
TF
|
TF:M01388_1
|
Factor: Dlx-5; motif: NNRGYAATTRNYKNNN; match class: 1
|
0.000894947159448298
|
5764
|
74
|
40
|
0.540540540540541
|
0.00693962526023595
|
|
TF
|
TF:M01275
|
Factor: ipf1; motif: CATTAR
|
0.000894947159448298
|
8839
|
93
|
63
|
0.67741935483871
|
0.00712750311121168
|
|
TF
|
TF:M11043
|
Factor: HMBOX1; motif: NCTAGTTAN
|
0.00109776476683506
|
597
|
58
|
10
|
0.172413793103448
|
0.016750418760469
|
|
TF
|
TF:M10650_1
|
Factor: HOX11L2; motif: NAATTGNNNNNNNNNNNNNCAATTN; match class: 1
|
0.00121287407268782
|
199
|
29
|
5
|
0.172413793103448
|
0.0251256281407035
|
|
TF
|
TF:M07988
|
Factor: EMX1; motif: NNTAATKANN
|
0.00150427550353765
|
3512
|
124
|
42
|
0.338709677419355
|
0.0119589977220957
|
|
TF
|
TF:M10696_1
|
Factor: HOXA6; motif: NGYMATTANN; match class: 1
|
0.00150427550353765
|
1223
|
38
|
11
|
0.289473684210526
|
0.00899427636958299
|
|
TF
|
TF:M08020_1
|
Factor: YY2; motif: NNCCGCCATTW; match class: 1
|
0.00150427550353765
|
72
|
141
|
6
|
0.0425531914893617
|
0.0833333333333333
|
|
TF
|
TF:M10695
|
Factor: HOXA6; motif: RTCRTTAN
|
0.0015127125063049
|
6486
|
89
|
49
|
0.550561797752809
|
0.00755473327166204
|
|
TF
|
TF:M10795_1
|
Factor: HOXA10; motif: NGTCGTAAAAN; match class: 1
|
0.0015127125063049
|
1609
|
25
|
10
|
0.4
|
0.00621504039776259
|
|
TF
|
TF:M02085
|
Factor: Bcl-6; motif: NTTYCTAGRA
|
0.0015301219379722
|
4976
|
17
|
13
|
0.764705882352941
|
0.00261254019292604
|
|
TF
|
TF:M04324_1
|
Factor: GBX2; motif: NNYAATTANN; match class: 1
|
0.00156924207633773
|
479
|
3
|
3
|
1
|
0.00626304801670146
|
|
TF
|
TF:M07258
|
Factor: HOXD10; motif: ARGAATAAWAA
|
0.00165259316540856
|
849
|
9
|
5
|
0.555555555555556
|
0.00588928150765607
|
|
TF
|
TF:M11620
|
Factor: Sox-14; motif: CCGAACAATN
|
0.00165259316540856
|
538
|
13
|
5
|
0.384615384615385
|
0.00929368029739777
|
|
TF
|
TF:M08982_1
|
Factor: YY1; motif: NAANATGGCGNNN; match class: 1
|
0.00165259316540856
|
1662
|
119
|
25
|
0.210084033613445
|
0.0150421179302046
|
|
TF
|
TF:M01388
|
Factor: Dlx-5; motif: NNRGYAATTRNYKNNN
|
0.00165259316540856
|
7668
|
74
|
47
|
0.635135135135135
|
0.00612936880542514
|
|
TF
|
TF:M10638_1
|
Factor: Nkx2-3; motif: NNCGTTRWS; match class: 1
|
0.00165259316540856
|
4099
|
109
|
42
|
0.385321100917431
|
0.0102464015613564
|
|
TF
|
TF:M12275_1
|
Factor: YY1; motif: NGCCGCCATYTTGN; match class: 1
|
0.00165324271839702
|
1600
|
116
|
24
|
0.206896551724138
|
0.015
|
|
TF
|
TF:M04328
|
Factor: GSX2; motif: NNYMATTANN
|
0.00165726946584114
|
6114
|
124
|
61
|
0.491935483870968
|
0.00997710173372588
|
|
TF
|
TF:M12296
|
Factor: ZNF282; motif: NTTCCCAYNACMCK
|
0.00168668582039051
|
343
|
10
|
4
|
0.4
|
0.0116618075801749
|
|
TF
|
TF:M10956_1
|
Factor: isx; motif: NTCRTTAA; match class: 1
|
0.00178349830399732
|
3336
|
26
|
14
|
0.538461538461538
|
0.00419664268585132
|
|
TF
|
TF:M11005_1
|
Factor: LHX4; motif: NNCRTTAN; match class: 1
|
0.00178349830399732
|
4203
|
114
|
44
|
0.385964912280702
|
0.0104687128241732
|
|
TF
|
TF:M10632
|
Factor: Nkx2-8; motif: STCGTTGAN
|
0.00178349830399732
|
4871
|
112
|
48
|
0.428571428571429
|
0.00985423937589817
|
|
TF
|
TF:M08261
|
Factor: ER71:HOXB13; motif: NNCGGAWGTNGTWAAN
|
0.00202244609099878
|
319
|
53
|
7
|
0.132075471698113
|
0.0219435736677116
|
|
TF
|
TF:M08282
|
Factor: Fli-1:Prrxl1; motif: ACCGGAAGTAATTAN
|
0.00215649695762934
|
2188
|
42
|
15
|
0.357142857142857
|
0.00685557586837294
|
|
TF
|
TF:M10685_1
|
Factor: HOXB7; motif: NGYAATTANN; match class: 1
|
0.00227987974388685
|
728
|
11
|
5
|
0.454545454545455
|
0.00686813186813187
|
|
TF
|
TF:M01235_1
|
Factor: ipf1; motif: NVSTAATTAC; match class: 1
|
0.00253882822268035
|
1569
|
64
|
16
|
0.25
|
0.0101975780752071
|
|
TF
|
TF:M01321
|
Factor: HOXC-8; motif: NNNNNGTAATTANNNT
|
0.00254581753941016
|
5641
|
74
|
38
|
0.513513513513513
|
0.00673639425633753
|
|
TF
|
TF:M10750
|
Factor: gsh-1; motif: NTMATNRN
|
0.00258636691170187
|
8022
|
124
|
73
|
0.588709677419355
|
0.00909997506856146
|
|
TF
|
TF:M04450_1
|
Factor: VSX1; motif: NGCYAATTRNN; match class: 1
|
0.00272867734309095
|
3267
|
34
|
16
|
0.470588235294118
|
0.00489745944291399
|
|
TF
|
TF:M04328_1
|
Factor: GSX2; motif: NNYMATTANN; match class: 1
|
0.00285816631432985
|
3836
|
119
|
42
|
0.352941176470588
|
0.0109489051094891
|
|
TF
|
TF:M01082_1
|
Factor: BRCA1:USF2; motif: KTNNGTTG; match class: 1
|
0.00290394795380171
|
1813
|
108
|
24
|
0.222222222222222
|
0.0132377275234418
|
|
TF
|
TF:M04616_1
|
Factor: islet1; motif: NGNTAATG; match class: 1
|
0.00294631952201782
|
59
|
22
|
3
|
0.136363636363636
|
0.0508474576271186
|
|
TF
|
TF:M11438_1
|
Factor: SAP-1; motif: NTCGTAAATGCN; match class: 1
|
0.00295710802089584
|
1191
|
8
|
5
|
0.625
|
0.00419815281276238
|
|
TF
|
TF:M08982
|
Factor: YY1; motif: NAANATGGCGNNN
|
0.00295710802089584
|
7125
|
100
|
56
|
0.56
|
0.00785964912280702
|
|
TF
|
TF:M10635
|
Factor: Nkx2-3; motif: GNCGTTRAG
|
0.00299075842633929
|
4594
|
108
|
44
|
0.407407407407407
|
0.00957771005659556
|
|
TF
|
TF:M10744
|
Factor: ipf1; motif: YTAATKRC
|
0.00305460404549137
|
1605
|
71
|
17
|
0.23943661971831
|
0.0105919003115265
|
|
TF
|
TF:M00489_1
|
Factor: Nkx6-2; motif: NWADTAAWTANN; match class: 1
|
0.00305460404549137
|
320
|
5
|
3
|
0.6
|
0.009375
|
|
TF
|
TF:M12127
|
Factor: ZNF396; motif: CTGTCCRAAAA
|
0.00308736077140679
|
142
|
10
|
3
|
0.3
|
0.0211267605633803
|
|
TF
|
TF:M07428
|
Factor: Six-3; motif: NWBTAATNYYWN
|
0.00313012962313416
|
3352
|
13
|
9
|
0.692307692307692
|
0.00268496420047733
|
|
TF
|
TF:M10736_1
|
Factor: HOXA2; motif: RTCRTTAR; match class: 1
|
0.00314814958150248
|
144
|
10
|
3
|
0.3
|
0.0208333333333333
|
|
TF
|
TF:M11507
|
Factor: CDP; motif: NTATTGATYR
|
0.00315512909321403
|
3157
|
11
|
8
|
0.727272727272727
|
0.00253405131453912
|
|
TF
|
TF:M07258_1
|
Factor: HOXD10; motif: ARGAATAAWAA; match class: 1
|
0.00333571660259491
|
56
|
25
|
3
|
0.12
|
0.0535714285714286
|
|
TF
|
TF:M10650
|
Factor: HOX11L2; motif: NAATTGNNNNNNNNNNNNNCAATTN
|
0.00339399752354142
|
339
|
5
|
3
|
0.6
|
0.00884955752212389
|
|
TF
|
TF:M04318
|
Factor: EN2; motif: NNYAATTANN
|
0.0034002491138712
|
1982
|
38
|
13
|
0.342105263157895
|
0.0065590312815338
|
|
TF
|
TF:M12286_1
|
Factor: ZNF647; motif: NYAGGCCYAC; match class: 1
|
0.0034122932816649
|
1636
|
10
|
6
|
0.6
|
0.00366748166259169
|
|
TF
|
TF:M11566
|
Factor: HNF-3alpha; motif: NWRWGTMAAYAW
|
0.0034122932816649
|
3516
|
64
|
25
|
0.390625
|
0.0071103526734926
|
|
TF
|
TF:M00483
|
Factor: ATF6; motif: TGACGTGG
|
0.00341908830936865
|
2627
|
113
|
31
|
0.274336283185841
|
0.0118005329272935
|
|
TF
|
TF:M12345_1
|
Factor: Zbtb37; motif: NYACCGCRNTCACCGCR; match class: 1
|
0.00364164393130398
|
1688
|
141
|
27
|
0.191489361702128
|
0.0159952606635071
|
|
TF
|
TF:M00430
|
Factor: E2F-1; motif: NTTSGCGG
|
0.00364164393130398
|
8016
|
142
|
81
|
0.570422535211268
|
0.0101047904191617
|
|
TF
|
TF:M00456_1
|
Factor: FAC1; motif: NNNCAMAACACRNA; match class: 1
|
0.00364164393130398
|
2699
|
91
|
27
|
0.296703296703297
|
0.0100037050759541
|
|
TF
|
TF:M00040_1
|
Factor: ATF-2; motif: TTACGTAA; match class: 1
|
0.00365455530599532
|
6460
|
63
|
36
|
0.571428571428571
|
0.00557275541795666
|
|
TF
|
TF:M04349_1
|
Factor: HOXB5; motif: NNYMATTANN; match class: 1
|
0.00375215596223088
|
4030
|
116
|
42
|
0.362068965517241
|
0.0104218362282878
|
|
TF
|
TF:M00023
|
Factor: HOXA5; motif: TGCNHNCWYCCYCATTAKTGNDCNMNHYCN
|
0.00379139444842096
|
3491
|
13
|
9
|
0.692307692307692
|
0.00257805786307648
|
|
TF
|
TF:M01275_1
|
Factor: ipf1; motif: CATTAR; match class: 1
|
0.00442106961812282
|
3429
|
86
|
30
|
0.348837209302326
|
0.00874890638670166
|
|
TF
|
TF:M10558_1
|
Factor: Prox-1; motif: NAAGGCGTCNTN; match class: 1
|
0.00452382810320312
|
91
|
18
|
3
|
0.166666666666667
|
0.032967032967033
|
|
TF
|
TF:M11331
|
Factor: Hlf; motif: NNTTACGTAANN
|
0.00473622559746198
|
543
|
66
|
9
|
0.136363636363636
|
0.0165745856353591
|
|
TF
|
TF:M02085_1
|
Factor: Bcl-6; motif: NTTYCTAGRA; match class: 1
|
0.00490770576396147
|
1072
|
21
|
7
|
0.333333333333333
|
0.00652985074626866
|
|
TF
|
TF:M10849_1
|
Factor: Cdx-2; motif: NRTCGTAANNNN; match class: 1
|
0.00513556023121428
|
4497
|
18
|
12
|
0.666666666666667
|
0.00266844563042028
|
|
TF
|
TF:M10953
|
Factor: isx; motif: NTCRTTAA
|
0.00513556023121428
|
5521
|
89
|
42
|
0.471910112359551
|
0.00760731751494294
|
|
TF
|
TF:M02026
|
Factor: MEF-2D; motif: WAAATAR
|
0.00513556023121428
|
7637
|
52
|
34
|
0.653846153846154
|
0.00445200995155166
|
|
TF
|
TF:M01385
|
Factor: Pax-4; motif: WNNNYTAATTARYNSNN
|
0.00513556023121428
|
3297
|
124
|
38
|
0.306451612903226
|
0.0115256293600243
|
|
TF
|
TF:M10053
|
Factor: AML3; motif: KGKNTGTGGTTTKK
|
0.00513556023121428
|
1332
|
65
|
14
|
0.215384615384615
|
0.0105105105105105
|
|
TF
|
TF:M11999_1
|
Factor: AML3; motif: NWAACCACRAAAACCACRAN; match class: 1
|
0.00513556023121428
|
1734
|
141
|
27
|
0.191489361702128
|
0.0155709342560554
|
|
TF
|
TF:M04318_1
|
Factor: EN2; motif: NNYAATTANN; match class: 1
|
0.00525731459565505
|
139
|
34
|
4
|
0.117647058823529
|
0.0287769784172662
|
|
TF
|
TF:M10703_1
|
Factor: HoxA5; motif: NYAATTAN; match class: 1
|
0.00563320096407906
|
2128
|
117
|
27
|
0.230769230769231
|
0.012687969924812
|
|
TF
|
TF:M04340_1
|
Factor: HOXA10; motif: NGYMATAAAAN; match class: 1
|
0.00584962753895158
|
193
|
10
|
3
|
0.3
|
0.0155440414507772
|
|
TF
|
TF:M04447
|
Factor: VAX2; motif: YTAATTAN
|
0.00585220124674014
|
2473
|
124
|
31
|
0.25
|
0.0125353821269713
|
|
TF
|
TF:M10869
|
Factor: Prrxl1; motif: YYAATTAN
|
0.00585220124674014
|
2473
|
124
|
31
|
0.25
|
0.0125353821269713
|
|
TF
|
TF:M10871
|
Factor: ARX; motif: NYAATTAN
|
0.00585220124674014
|
2473
|
124
|
31
|
0.25
|
0.0125353821269713
|
|
TF
|
TF:M07428_1
|
Factor: Six-3; motif: NWBTAATNYYWN; match class: 1
|
0.00585220124674014
|
596
|
4
|
3
|
0.75
|
0.00503355704697987
|
|
TF
|
TF:M11975_1
|
Factor: RFX7; motif: SGTTGCTRN; match class: 1
|
0.00587171537569998
|
186
|
115
|
7
|
0.0608695652173913
|
0.0376344086021505
|
|
TF
|
TF:M00076
|
Factor: GATA-2; motif: NNNGATRNNN
|
0.00607906039657468
|
3216
|
5
|
5
|
1
|
0.0015547263681592
|
|
TF
|
TF:M01255
|
Factor: ipf1; motif: RNWCATTAANWN
|
0.00615326455317118
|
3962
|
88
|
33
|
0.375
|
0.00832912670368501
|
|
TF
|
TF:M01362_1
|
Factor: CART1; motif: NGNNYTAATTARTNNNN; match class: 1
|
0.00626244078325473
|
3461
|
116
|
37
|
0.318965517241379
|
0.0106905518636232
|
|
TF
|
TF:M01035
|
Factor: YY1; motif: NYNKCCATNTT
|
0.00628911378999489
|
5924
|
116
|
54
|
0.46551724137931
|
0.00911546252532073
|
|
TF
|
TF:M12049
|
Factor: znf580; motif: NCTACCCYNNNMCTACCCT
|
0.00628911378999489
|
77
|
25
|
3
|
0.12
|
0.038961038961039
|
|
TF
|
TF:M12273
|
Factor: YY1; motif: GGCSGCCATTTTGN
|
0.00628911378999489
|
6229
|
116
|
56
|
0.482758620689655
|
0.00899020709584203
|
|
TF
|
TF:M11725
|
Factor: B-Myb; motif: NTAACSGTYRN
|
0.00628911378999489
|
863
|
66
|
11
|
0.166666666666667
|
0.0127462340672074
|
|
TF
|
TF:M10765_1
|
Factor: Meox2; motif: NTCRTTAN; match class: 1
|
0.00628911378999489
|
206
|
10
|
3
|
0.3
|
0.0145631067961165
|
|
TF
|
TF:M10733_1
|
Factor: HOXA2; motif: RTCRTTAN; match class: 1
|
0.00628911378999489
|
206
|
10
|
3
|
0.3
|
0.0145631067961165
|
|
TF
|
TF:M07375
|
Factor: BCL-6; motif: TTCCTAGAAA
|
0.00628911378999489
|
556
|
84
|
10
|
0.119047619047619
|
0.0179856115107914
|
|
TF
|
TF:M11037_1
|
Factor: Six-6; motif: NNSTATCRNN; match class: 1
|
0.00631292992789442
|
938
|
18
|
6
|
0.333333333333333
|
0.00639658848614072
|
|
TF
|
TF:M04344
|
Factor: HOXA2; motif: NNTMATTANN
|
0.00653403467407844
|
2557
|
95
|
26
|
0.273684210526316
|
0.0101681658193195
|
|
TF
|
TF:M12275
|
Factor: YY1; motif: NGCCGCCATYTTGN
|
0.00681052900056439
|
7043
|
116
|
61
|
0.525862068965517
|
0.00866108192531592
|
|
TF
|
TF:M04332
|
Factor: HMBOX1; motif: MYTAGTTAMN
|
0.00681052900056439
|
1501
|
17
|
7
|
0.411764705882353
|
0.00466355762824783
|
|
TF
|
TF:M02044
|
Factor: YY1; motif: GCCGCCATTTTG
|
0.00681052900056439
|
6102
|
116
|
55
|
0.474137931034483
|
0.00901343821697804
|
|
TF
|
TF:M01464_1
|
Factor: HOXA10; motif: NNNGYAATAAAATNNW; match class: 1
|
0.00683555355398845
|
529
|
11
|
4
|
0.363636363636364
|
0.00756143667296786
|
|
TF
|
TF:M03924_1
|
Factor: YY1; motif: NNCGCCATTNN; match class: 1
|
0.00683555355398845
|
3700
|
142
|
45
|
0.316901408450704
|
0.0121621621621622
|
|
TF
|
TF:M01712_1
|
Factor: CRX; motif: YTAATC; match class: 1
|
0.00683555355398845
|
5021
|
11
|
9
|
0.818181818181818
|
0.00179247161919936
|
|
TF
|
TF:M00489
|
Factor: Nkx6-2; motif: NWADTAAWTANN
|
0.00683905862536993
|
1779
|
71
|
17
|
0.23943661971831
|
0.00955593029792018
|
|
TF
|
TF:M12295
|
Factor: ZNF282; motif: NTTCCCAYAACMCN
|
0.00690612680627393
|
668
|
4
|
3
|
0.75
|
0.00449101796407186
|
|
TF
|
TF:M01759_1
|
Factor: AML2; motif: AACCRCAA; match class: 1
|
0.00690612680627393
|
237
|
67
|
6
|
0.0895522388059701
|
0.0253164556962025
|
|
TF
|
TF:M11956
|
Factor: POU6F1; motif: TMATTANNNCTCATTA
|
0.00692602959118593
|
284
|
80
|
7
|
0.0875
|
0.0246478873239437
|
|
TF
|
TF:M12309
|
Factor: ZNF597; motif: GGCGGCCATYTTGN
|
0.00708386327109715
|
64
|
84
|
4
|
0.0476190476190476
|
0.0625
|
|
TF
|
TF:M10702_1
|
Factor: HOXB5; motif: STAATTAS; match class: 1
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M01082
|
Factor: BRCA1:USF2; motif: KTNNGTTG
|
0.00722371433337181
|
7155
|
30
|
21
|
0.7
|
0.00293501048218029
|
|
TF
|
TF:M10740
|
Factor: HOXB2; motif: NYAATTAN
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M10729
|
Factor: HOXC-8; motif: RTCRTTAN
|
0.00722371433337181
|
3956
|
13
|
9
|
0.692307692307692
|
0.00227502527805865
|
|
TF
|
TF:M10868
|
Factor: EVX2; motif: NTAATTAN
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M10756
|
Factor: hoxd1; motif: NTAATTAN
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M10756_1
|
Factor: hoxd1; motif: NTAATTAN; match class: 1
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M10751_1
|
Factor: HOXA1; motif: NTAATTAN; match class: 1
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M10723
|
Factor: HOXD8; motif: RTCRTTAN
|
0.00722371433337181
|
4249
|
25
|
14
|
0.56
|
0.00329489291598023
|
|
TF
|
TF:M10868_1
|
Factor: EVX2; motif: NTAATTAN; match class: 1
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M08772_1
|
Factor: HOXA6; motif: NYMATTAN; match class: 1
|
0.00722371433337181
|
2568
|
38
|
14
|
0.368421052631579
|
0.00545171339563863
|
|
TF
|
TF:M10700
|
Factor: HOXB5; motif: NTAATTAN
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M10751
|
Factor: HOXA1; motif: NTAATTAN
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M10700_1
|
Factor: HOXB5; motif: NTAATTAN; match class: 1
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M10702
|
Factor: HOXB5; motif: STAATTAS
|
0.00722371433337181
|
1111
|
102
|
16
|
0.156862745098039
|
0.0144014401440144
|
|
TF
|
TF:M01013_1
|
Factor: ipf1; motif: TSNGYCATTANNNNC; match class: 1
|
0.00723653949695385
|
232
|
10
|
3
|
0.3
|
0.0129310344827586
|
|
TF
|
TF:M07304
|
Factor: Pbx; motif: NTGATTGANN
|
0.00734208527654509
|
1571
|
5
|
4
|
0.8
|
0.00254614894971356
|
|
TF
|
TF:M10526_1
|
Factor: Blimp-1; motif: RRGNGAAAGNRANNN; match class: 1
|
0.00734208527654509
|
286
|
58
|
6
|
0.103448275862069
|
0.020979020979021
|
|
TF
|
TF:M08785
|
Factor: Sox-6; motif: CACCGAACAAT
|
0.00734208527654509
|
1105
|
23
|
7
|
0.304347826086957
|
0.00633484162895928
|
|
TF
|
TF:M01321_1
|
Factor: HOXC-8; motif: NNNNNGTAATTANNNT; match class: 1
|
0.00734208527654509
|
4392
|
119
|
44
|
0.369747899159664
|
0.0100182149362477
|
|
TF
|
TF:M04279
|
Factor: ALX3; motif: NNYAATTANN
|
0.00734208527654509
|
3934
|
124
|
42
|
0.338709677419355
|
0.0106761565836299
|
|
TF
|
TF:M04814
|
Factor: RelA-p65; motif: AAATCCCCT
|
0.00735650928411866
|
4528
|
52
|
24
|
0.461538461538462
|
0.00530035335689046
|
|
TF
|
TF:M01233
|
Factor: ipf1; motif: MVCTAATTAS
|
0.00770237355440885
|
1554
|
122
|
22
|
0.180327868852459
|
0.0141570141570142
|
|
TF
|
TF:M08930
|
Factor: JUND:FRA-1; motif: RTGACGTMAY
|
0.00780911110742259
|
5702
|
29
|
18
|
0.620689655172414
|
0.00315678709224833
|
|
TF
|
TF:M10718
|
Factor: HOXC4; motif: RTCRTTAN
|
0.00787845447145843
|
2772
|
11
|
7
|
0.636363636363636
|
0.00252525252525253
|
|
TF
|
TF:M00126
|
Factor: GATA-1; motif: NNNNNGATANKGGN
|
0.00787845447145843
|
3887
|
11
|
8
|
0.727272727272727
|
0.00205814252636995
|
|
TF
|
TF:M04450
|
Factor: VSX1; motif: NGCYAATTRNN
|
0.00798338750674626
|
7084
|
72
|
41
|
0.569444444444444
|
0.00578769057029927
|
|
TF
|
TF:M04614
|
Factor: Ipf1; motif: NNNNTAATKR
|
0.00798338750674626
|
1673
|
122
|
23
|
0.188524590163934
|
0.013747758517633
|
|
TF
|
TF:M00425_1
|
Factor: E2F; motif: TTTCGCGC; match class: 1
|
0.00798338750674626
|
5355
|
135
|
56
|
0.414814814814815
|
0.0104575163398693
|
|
TF
|
TF:M12285
|
Factor: ZNF647; motif: NTAGGCCTAN
|
0.00801345821412085
|
1290
|
10
|
5
|
0.5
|
0.00387596899224806
|
|
TF
|
TF:M07234
|
Factor: YY1; motif: CAARATGGCNGC
|
0.00801345821412085
|
6045
|
116
|
54
|
0.46551724137931
|
0.00893300248138958
|
|
TF
|
TF:M10965
|
Factor: ESX1L; motif: NYAATTAN
|
0.0082733933458919
|
3236
|
25
|
12
|
0.48
|
0.00370828182941904
|
|
TF
|
TF:M11225_1
|
Factor: CREM; motif: NNTGACGTCANN; match class: 1
|
0.00827935169208795
|
2482
|
140
|
33
|
0.235714285714286
|
0.0132957292506044
|
|
TF
|
TF:M11223
|
Factor: CREB1; motif: NRTGACGTCANN
|
0.00827935169208795
|
2810
|
140
|
36
|
0.257142857142857
|
0.0128113879003559
|
|
TF
|
TF:M03560_1
|
Factor: PMX1; motif: TAATHA; match class: 1
|
0.00827935169208795
|
10273
|
76
|
55
|
0.723684210526316
|
0.00535384016353548
|
|
TF
|
TF:M12005_1
|
Factor: GMEB-1; motif: KNACGTNRNNACGTAN; match class: 1
|
0.00832688578629859
|
471
|
107
|
10
|
0.0934579439252336
|
0.0212314225053079
|
|
TF
|
TF:M10690_1
|
Factor: HOXA7; motif: GYMATTAN; match class: 1
|
0.00832688578629859
|
6126
|
67
|
35
|
0.522388059701492
|
0.00571335292197192
|
|
TF
|
TF:M10615_1
|
Factor: Dlx-5; motif: NTCRTTAN; match class: 1
|
0.00832688578629859
|
730
|
25
|
6
|
0.24
|
0.00821917808219178
|
|
TF
|
TF:M10753_1
|
Factor: HOXA1; motif: NTAATTAN; match class: 1
|
0.00844469452874124
|
1144
|
102
|
16
|
0.156862745098039
|
0.013986013986014
|
|
TF
|
TF:M10754_1
|
Factor: hoxd1; motif: NTAATTAS; match class: 1
|
0.00844469452874124
|
1144
|
102
|
16
|
0.156862745098039
|
0.013986013986014
|
|
TF
|
TF:M10104
|
Factor: YY1; motif: CAARATGGCGGC
|
0.00848225591756818
|
6071
|
116
|
54
|
0.46551724137931
|
0.00889474551144787
|
|
TF
|
TF:M10706_1
|
Factor: HoxA5; motif: NYMATTAN; match class: 1
|
0.00851994016983053
|
2836
|
124
|
33
|
0.266129032258065
|
0.0116361071932299
|
|
TF
|
TF:M08232
|
Factor: Elk-1:TEF; motif: RCCGGAAGTTACGTAAN
|
0.00853152088680138
|
6188
|
69
|
36
|
0.521739130434783
|
0.00581771170006464
|
|
TF
|
TF:M07327
|
Factor: Nanog; motif: NTNTAATGSN
|
0.00853152088680138
|
1926
|
88
|
20
|
0.227272727272727
|
0.0103842159916926
|
|
TF
|
TF:M00040
|
Factor: ATF-2; motif: TTACGTAA
|
0.00866580643021977
|
6902
|
63
|
36
|
0.571428571428571
|
0.00521587945523037
|
|
TF
|
TF:M10704_1
|
Factor: HoxA5; motif: RTCATTAN; match class: 1
|
0.00867242014304678
|
1186
|
11
|
5
|
0.454545454545455
|
0.00421585160202361
|
|
TF
|
TF:M01449_1
|
Factor: Cdx-2; motif: NNNGGYMATAAAANNN; match class: 1
|
0.008726275341562
|
265
|
10
|
3
|
0.3
|
0.0113207547169811
|
|
TF
|
TF:M04446
|
Factor: VAX1; motif: YTAATTAN
|
0.00878081831225494
|
3224
|
124
|
36
|
0.290322580645161
|
0.011166253101737
|
|
TF
|
TF:M04299
|
Factor: Dlx-1; motif: NNTAATTANN
|
0.00878081831225494
|
2524
|
122
|
30
|
0.245901639344262
|
0.0118858954041204
|
|
TF
|
TF:M10673
|
Factor: Emx-2; motif: YTAATTAN
|
0.00878081831225494
|
3224
|
124
|
36
|
0.290322580645161
|
0.011166253101737
|
|
TF
|
TF:M12273_1
|
Factor: YY1; motif: GGCSGCCATTTTGN; match class: 1
|
0.00878081831225494
|
1261
|
142
|
21
|
0.147887323943662
|
0.0166534496431404
|
|
TF
|
TF:M01171
|
Factor: BCL6; motif: NKTTCTAGGW
|
0.00878081831225494
|
1344
|
10
|
5
|
0.5
|
0.0037202380952381
|
|
TF
|
TF:M01425_1
|
Factor: HNF-1beta; motif: NNNNGTTAACTAGNNNT; match class: 1
|
0.00880062843823344
|
1737
|
5
|
4
|
0.8
|
0.0023028209556707
|
|
TF
|
TF:M03929_1
|
Factor: ZBTB7C; motif: NCGACCACCGNN; match class: 1
|
0.0088211744178478
|
303
|
109
|
8
|
0.073394495412844
|
0.0264026402640264
|
|
TF
|
TF:M10736
|
Factor: HOXA2; motif: RTCRTTAR
|
0.00883923924545108
|
1961
|
11
|
6
|
0.545454545454545
|
0.00305966343702193
|
|
TF
|
TF:M11515
|
Factor: DMRT1; motif: NNNNGCTACATN
|
0.00883923924545108
|
3857
|
5
|
5
|
1
|
0.00129634430904848
|
|
TF
|
TF:M00793
|
Factor: YY1; motif: GCCATNTTN
|
0.00883923924545108
|
5186
|
116
|
48
|
0.413793103448276
|
0.00925568839182414
|
|
TF
|
TF:M07370
|
Factor: YY1; motif: NNNNAARATGGNNNN
|
0.00898172993852987
|
5811
|
124
|
55
|
0.443548387096774
|
0.00946480812252624
|
|
TF
|
TF:M01438
|
Factor: ipf1; motif: NNRNTAATTAGYNCAN
|
0.00898172993852987
|
3325
|
74
|
25
|
0.337837837837838
|
0.0075187969924812
|
|
TF
|
TF:M10706
|
Factor: HoxA5; motif: NYMATTAN
|
0.00898172993852987
|
3532
|
74
|
26
|
0.351351351351351
|
0.00736126840317101
|
|
TF
|
TF:M10705_1
|
Factor: HoxA5; motif: GYMATTAS; match class: 1
|
0.00898172993852987
|
3532
|
74
|
26
|
0.351351351351351
|
0.00736126840317101
|
|
TF
|
TF:M10705
|
Factor: HoxA5; motif: GYMATTAS
|
0.00898172993852987
|
3532
|
74
|
26
|
0.351351351351351
|
0.00736126840317101
|
|
TF
|
TF:M10703
|
Factor: HoxA5; motif: NYAATTAN
|
0.00898172993852987
|
3532
|
74
|
26
|
0.351351351351351
|
0.00736126840317101
|
|
TF
|
TF:M11438
|
Factor: SAP-1; motif: NTCGTAAATGCN
|
0.00917282720534854
|
5901
|
109
|
50
|
0.458715596330275
|
0.00847314014573801
|
|
TF
|
TF:M10826
|
Factor: HOXC12; motif: NGTAATAAAAN
|
0.00919877404992019
|
721
|
10
|
4
|
0.4
|
0.00554785020804438
|
|
TF
|
TF:M11904_1
|
Factor: POU3F2; motif: WTATGCWAATKAG; match class: 1
|
0.00919877404992019
|
430
|
28
|
5
|
0.178571428571429
|
0.0116279069767442
|
|
TF
|
TF:M01431_1
|
Factor: Barx-2; motif: TAANTAATTANTKNTN; match class: 1
|
0.00920780228822326
|
1466
|
25
|
8
|
0.32
|
0.00545702592087312
|
|
TF
|
TF:M10692_1
|
Factor: HOXB6; motif: GTCGTTAN; match class: 1
|
0.0092247204186171
|
1009
|
13
|
5
|
0.384615384615385
|
0.00495540138751239
|
|
TF
|
TF:M08930_1
|
Factor: JUND:FRA-1; motif: RTGACGTMAY; match class: 1
|
0.00937186814038585
|
1607
|
122
|
22
|
0.180327868852459
|
0.0136901057871811
|
|
TF
|
TF:M10718_1
|
Factor: HOXC4; motif: RTCRTTAN; match class: 1
|
0.00938825267250075
|
283
|
10
|
3
|
0.3
|
0.0106007067137809
|
|
TF
|
TF:M10739_1
|
Factor: HOXB2; motif: RTCATTAN; match class: 1
|
0.00938825267250075
|
653
|
11
|
4
|
0.363636363636364
|
0.00612557427258806
|
|
TF
|
TF:M08392
|
Factor: A-Myb:TBR2; motif: NNGTGNNNAACGG
|
0.00972289632357282
|
2093
|
108
|
24
|
0.222222222222222
|
0.0114667940754897
|
|
TF
|
TF:M10667_1
|
Factor: NOTO; motif: NYMATTAN; match class: 1
|
0.0100748890524743
|
2236
|
67
|
18
|
0.26865671641791
|
0.00805008944543828
|
|
TF
|
TF:M01412
|
Factor: Msx-1; motif: NNNNANTAATTANTNN
|
0.0101926241542049
|
3098
|
71
|
23
|
0.323943661971831
|
0.00742414460942544
|
|
TF
|
TF:M01658_1
|
Factor: AML1; motif: TGTGGTK; match class: 1
|
0.0104171882617565
|
625
|
31
|
6
|
0.193548387096774
|
0.0096
|
|
TF
|
TF:M10948
|
Factor: rax; motif: NNCRTTAN
|
0.0104171882617565
|
11816
|
95
|
73
|
0.768421052631579
|
0.00617806364251862
|
|
TF
|
TF:M11218_1
|
Factor: CREB1; motif: NRTGAYGCGTN; match class: 1
|
0.010625604645697
|
2438
|
13
|
7
|
0.538461538461538
|
0.00287120590648072
|
|
TF
|
TF:M04324
|
Factor: GBX2; motif: NNYAATTANN
|
0.0107647908252664
|
3485
|
10
|
7
|
0.7
|
0.00200860832137733
|
|
TF
|
TF:M11668
|
Factor: IRF-6; motif: ACCGATACC
|
0.0108487390761
|
406
|
86
|
8
|
0.0930232558139535
|
0.0197044334975369
|
|
TF
|
TF:M07458
|
Factor: HOXC13; motif: CTMATTAAANNRKT
|
0.0109874403793163
|
259
|
27
|
4
|
0.148148148148148
|
0.0154440154440154
|
|
TF
|
TF:M10727
|
Factor: HOXB8; motif: RTCRTTAN
|
0.0109874403793163
|
6289
|
79
|
40
|
0.506329113924051
|
0.00636031165527111
|
|
TF
|
TF:M01020_1
|
Factor: Tbx5; motif: TNAGGTGTKV; match class: 1
|
0.0110444677256478
|
4140
|
5
|
5
|
1
|
0.00120772946859903
|
|
TF
|
TF:M11724_1
|
Factor: B-Myb; motif: NTRACSGTYNN; match class: 1
|
0.0110444677256478
|
1158
|
66
|
12
|
0.181818181818182
|
0.0103626943005181
|
|
TF
|
TF:M11724
|
Factor: B-Myb; motif: NTRACSGTYNN
|
0.0110444677256478
|
1158
|
66
|
12
|
0.181818181818182
|
0.0103626943005181
|
|
TF
|
TF:M01104
|
Factor: MOVO-B; motif: GNGGGGG
|
0.0110550843717897
|
10261
|
105
|
72
|
0.685714285714286
|
0.00701685995517006
|
|
TF
|
TF:M00210_1
|
Factor: OCT-x; motif: CTNATTTGCATAY; match class: 1
|
0.0112919396181019
|
700
|
5
|
3
|
0.6
|
0.00428571428571429
|
|
TF
|
TF:M10436_1
|
Factor: YY1; motif: CAANATGGCGGC; match class: 1
|
0.0115525571863061
|
2285
|
119
|
27
|
0.226890756302521
|
0.0118161925601751
|
|
TF
|
TF:M10806
|
Factor: HOXC10; motif: NGYMATAAAAN
|
0.0115657630824277
|
787
|
10
|
4
|
0.4
|
0.00508259212198221
|
|
TF
|
TF:M11223_1
|
Factor: CREB1; motif: NRTGACGTCANN; match class: 1
|
0.0117095869165711
|
2678
|
140
|
34
|
0.242857142857143
|
0.0126960418222554
|
|
TF
|
TF:M04711
|
Factor: PU.1; motif: ACTTCCTCT
|
0.0117095869165711
|
1712
|
34
|
10
|
0.294117647058824
|
0.0058411214953271
|
|
TF
|
TF:M10108
|
Factor: WT1; motif: RGGNGGGGGAGGRGGNGGRG
|
0.0117659137377783
|
6511
|
116
|
56
|
0.482758620689655
|
0.00860082936568883
|
|
TF
|
TF:M11131
|
Factor: Ngn-2; motif: RACATATGTY
|
0.0117885677207704
|
900
|
9
|
4
|
0.444444444444444
|
0.00444444444444444
|
|
TF
|
TF:M10785
|
Factor: hoxa9; motif: RTCGTWANNN
|
0.011791625686606
|
5632
|
26
|
16
|
0.615384615384615
|
0.00284090909090909
|
|
TF
|
TF:M04748_1
|
Factor: GABP-alpha; motif: AACCGGAAR; match class: 1
|
0.011791625686606
|
10368
|
136
|
91
|
0.669117647058823
|
0.00877700617283951
|
|
TF
|
TF:M04399
|
Factor: MEOX2; motif: NSTAATTANN
|
0.011791625686606
|
3837
|
74
|
27
|
0.364864864864865
|
0.00703674745895231
|
|
TF
|
TF:M10761
|
Factor: MOX1; motif: NTCRTTAN
|
0.011791625686606
|
3158
|
46
|
17
|
0.369565217391304
|
0.00538315389487017
|
|
TF
|
TF:M00695
|
Factor: ETF; motif: GVGGMGG
|
0.011791625686606
|
10430
|
142
|
95
|
0.669014084507042
|
0.00910834132310642
|
|
TF
|
TF:M08771_1
|
Factor: HOXA4; motif: RYCATTRN; match class: 1
|
0.011791625686606
|
320
|
10
|
3
|
0.3
|
0.009375
|
|
TF
|
TF:M01306_1
|
Factor: ZABC1; motif: ATTCCNAC; match class: 1
|
0.011791625686606
|
2690
|
23
|
10
|
0.434782608695652
|
0.00371747211895911
|
|
TF
|
TF:M00930
|
Factor: Oct-1; motif: TNATTTGCATW
|
0.011791625686606
|
1972
|
5
|
4
|
0.8
|
0.00202839756592292
|
|
TF
|
TF:M00133
|
Factor: Tst-1; motif: NNKGAATTAVAVTDN
|
0.0118401506772095
|
5683
|
45
|
24
|
0.533333333333333
|
0.00422312159070913
|
|
TF
|
TF:M10745
|
Factor: ipf1; motif: RTCATTAN
|
0.0119997416273445
|
2179
|
94
|
22
|
0.234042553191489
|
0.0100963744837081
|
|
TF
|
TF:M04407
|
Factor: MSX2; motif: NCAATTAN
|
0.0121418222178094
|
736
|
5
|
3
|
0.6
|
0.00407608695652174
|
|
TF
|
TF:M10561
|
Factor: Msx-2; motif: NCAATTAN
|
0.0121418222178094
|
736
|
5
|
3
|
0.6
|
0.00407608695652174
|
|
TF
|
TF:M10436
|
Factor: YY1; motif: CAANATGGCGGC
|
0.012367650774314
|
8220
|
99
|
58
|
0.585858585858586
|
0.00705596107055961
|
|
TF
|
TF:M01408
|
Factor: Brn-3c; motif: NNTTATTAATKANKNC
|
0.0123685147384104
|
4888
|
60
|
27
|
0.45
|
0.00552373158756138
|
|
TF
|
TF:M03546_1
|
Factor: Dlx-5; motif: AATTAN; match class: 1
|
0.0124726403114727
|
5831
|
11
|
9
|
0.818181818181818
|
0.00154347453267021
|
|
TF
|
TF:M01654_1
|
Factor: DRI1; motif: AATTAA; match class: 1
|
0.0124726403114727
|
5831
|
11
|
9
|
0.818181818181818
|
0.00154347453267021
|
|
TF
|
TF:M10573
|
Factor: Gbx2; motif: NYAATTAN
|
0.0125668446210863
|
614
|
13
|
4
|
0.307692307692308
|
0.00651465798045603
|
|
TF
|
TF:M00039
|
Factor: CREB; motif: TGACGTMA
|
0.0125668446210863
|
5222
|
124
|
50
|
0.403225806451613
|
0.00957487552661815
|
|
TF
|
TF:M10574
|
Factor: Gbx2; motif: NYAATTAN
|
0.0125668446210863
|
614
|
13
|
4
|
0.307692307692308
|
0.00651465798045603
|
|
TF
|
TF:M10578_1
|
Factor: Barhl-1; motif: NTAAACGN; match class: 1
|
0.0125668446210863
|
1391
|
16
|
6
|
0.375
|
0.00431344356578001
|
|
TF
|
TF:M10571
|
Factor: Gbx2; motif: NYAATTAN
|
0.0125668446210863
|
614
|
13
|
4
|
0.307692307692308
|
0.00651465798045603
|
|
TF
|
TF:M01449
|
Factor: Cdx-2; motif: NNNGGYMATAAAANNN
|
0.0125668446210863
|
2485
|
10
|
6
|
0.6
|
0.00241448692152917
|
|
TF
|
TF:M04405
|
Factor: Msx-1; motif: NCAATTAN
|
0.0125668446210863
|
614
|
13
|
4
|
0.307692307692308
|
0.00651465798045603
|
|
TF
|
TF:M08242
|
Factor: ERF:HOXB13; motif: NNCGGAARYNRTWAAN
|
0.0125668446210863
|
754
|
5
|
3
|
0.6
|
0.00397877984084881
|
|
TF
|
TF:M10860
|
Factor: En-1; motif: NYAATTAN
|
0.0125668446210863
|
614
|
13
|
4
|
0.307692307692308
|
0.00651465798045603
|
|
TF
|
TF:M01017
|
Factor: Pbx-1; motif: NNATCAATCANN
|
0.0125830856909621
|
2306
|
36
|
12
|
0.333333333333333
|
0.00520381613183001
|
|
TF
|
TF:M10569
|
Factor: HB9; motif: NTCRTTAN
|
0.0126324827841514
|
3209
|
11
|
7
|
0.636363636363636
|
0.00218136491118729
|
|
TF
|
TF:M00228
|
Factor: VBP; motif: GTTACRTMAT
|
0.0126487614297589
|
2565
|
66
|
19
|
0.287878787878788
|
0.00740740740740741
|
|
TF
|
TF:M11313_1
|
Factor: CREBPA; motif: NRTGACGTMANN; match class: 1
|
0.0128936442653785
|
2025
|
76
|
18
|
0.236842105263158
|
0.00888888888888889
|
|
TF
|
TF:M10701_1
|
Factor: HOXB5; motif: GTCATTAN; match class: 1
|
0.0131854401411289
|
1396
|
11
|
5
|
0.454545454545455
|
0.00358166189111748
|
|
TF
|
TF:M08635
|
Factor: TEF-3:Dlx-3; motif: RCATWCCNNNYAATTA
|
0.0131854401411289
|
238
|
14
|
3
|
0.214285714285714
|
0.0126050420168067
|
|
TF
|
TF:M11309_1
|
Factor: ATF-2; motif: NRTGACGTMANN; match class: 1
|
0.0131854401411289
|
2293
|
27
|
10
|
0.37037037037037
|
0.00436109899694723
|
|
TF
|
TF:M10785_1
|
Factor: hoxa9; motif: RTCGTWANNN; match class: 1
|
0.0131854401411289
|
1160
|
59
|
11
|
0.186440677966102
|
0.00948275862068966
|
|
TF
|
TF:M11526_1
|
Factor: E2F-3; motif: NTTTTGGCGCCAAAAN; match class: 1
|
0.0133848462445943
|
6798
|
33
|
21
|
0.636363636363636
|
0.0030891438658429
|
|
TF
|
TF:M10816_1
|
Factor: HOXC11; motif: NGYMATWAANN; match class: 1
|
0.0133848462445943
|
1055
|
4
|
3
|
0.75
|
0.0028436018957346
|
|
TF
|
TF:M10692
|
Factor: HOXB6; motif: GTCGTTAN
|
0.0135581691657044
|
5380
|
25
|
15
|
0.6
|
0.00278810408921933
|
|
TF
|
TF:M01658
|
Factor: AML1; motif: TGTGGTK
|
0.0136331310931233
|
4477
|
5
|
5
|
1
|
0.00111681929863748
|
|
TF
|
TF:M03862
|
Factor: YB-1; motif: NNNNCCAATNN
|
0.0139067077212722
|
9198
|
69
|
46
|
0.666666666666667
|
0.00500108719286801
|
|
TF
|
TF:M04069_1
|
Factor: POU1F1; motif: AWTATGCWAATKAG; match class: 1
|
0.0141362251549199
|
501
|
28
|
5
|
0.178571428571429
|
0.00998003992015968
|
|
TF
|
TF:M11330
|
Factor: Hlf; motif: NGTTRYGYAANN
|
0.0145753289103765
|
1046
|
66
|
11
|
0.166666666666667
|
0.0105162523900574
|
|
TF
|
TF:M02099_1
|
Factor: MITF; motif: CATGTGM; match class: 1
|
0.0145834354323059
|
359
|
10
|
3
|
0.3
|
0.00835654596100279
|
|
TF
|
TF:M04748
|
Factor: GABP-alpha; motif: AACCGGAAR
|
0.0146243776525038
|
16027
|
109
|
101
|
0.926605504587156
|
0.00630186560179697
|
|
TF
|
TF:M10761_1
|
Factor: MOX1; motif: NTCRTTAN; match class: 1
|
0.0149109829109332
|
363
|
10
|
3
|
0.3
|
0.00826446280991736
|
|
TF
|
TF:M04696
|
Factor: YY1; motif: GCCGCCATNTTGNNNNNGGNCN
|
0.0149109829109332
|
7819
|
117
|
64
|
0.547008547008547
|
0.00818518992198491
|
|
TF
|
TF:M08482_1
|
Factor: FOXO1A:HOXA10; motif: RWMAACANCRTWAA; match class: 1
|
0.0150359911308593
|
702
|
31
|
6
|
0.193548387096774
|
0.00854700854700855
|
|
TF
|
TF:M11134_1
|
Factor: Ngn-2; motif: RACATATGTY; match class: 1
|
0.0158668540676717
|
1235
|
66
|
12
|
0.181818181818182
|
0.0097165991902834
|
|
TF
|
TF:M04616
|
Factor: islet1; motif: NGNTAATG
|
0.0160728643405891
|
1141
|
4
|
3
|
0.75
|
0.00262927256792287
|
|
TF
|
TF:M00917
|
Factor: CREB; motif: CNNTGACGTMA
|
0.0160728643405891
|
5578
|
124
|
52
|
0.419354838709677
|
0.00932233775546791
|
|
TF
|
TF:M10694
|
Factor: HOXA6; motif: NGYMATTANN
|
0.0162604586186134
|
3321
|
74
|
24
|
0.324324324324324
|
0.00722673893405601
|
|
TF
|
TF:M11309
|
Factor: ATF-2; motif: NRTGACGTMANN
|
0.0164095724715317
|
6429
|
77
|
39
|
0.506493506493506
|
0.00606626224918339
|
|
TF
|
TF:M10052
|
Factor: AML1; motif: NYTGTGGTTWN
|
0.0166596851575111
|
1086
|
65
|
11
|
0.169230769230769
|
0.0101289134438306
|
|
TF
|
TF:M11307
|
Factor: ATF-2; motif: NNTGACGTMANN
|
0.0167386356072456
|
2085
|
76
|
18
|
0.236842105263158
|
0.00863309352517986
|
|
TF
|
TF:M10776
|
Factor: hoxd9; motif: GYMATAAAAN
|
0.0168695594048713
|
921
|
10
|
4
|
0.4
|
0.00434310532030402
|
|
TF
|
TF:M11200
|
Factor: FIGLA; motif: NMCACCTGK
|
0.0170686695810534
|
386
|
10
|
3
|
0.3
|
0.0077720207253886
|
|
TF
|
TF:M11201
|
Factor: FIGLA; motif: NMCACCTGN
|
0.0170686695810534
|
386
|
10
|
3
|
0.3
|
0.0077720207253886
|
|
TF
|
TF:M10765
|
Factor: Meox2; motif: NTCRTTAN
|
0.0170915723165948
|
2373
|
11
|
6
|
0.545454545454545
|
0.00252844500632111
|
|
TF
|
TF:M10733
|
Factor: HOXA2; motif: RTCRTTAN
|
0.0170915723165948
|
2373
|
11
|
6
|
0.545454545454545
|
0.00252844500632111
|
|
TF
|
TF:M01325_1
|
Factor: LHX2; motif: WNNACTAATTAGNNNNN; match class: 1
|
0.0174007362617238
|
651
|
124
|
12
|
0.0967741935483871
|
0.0184331797235023
|
|
TF
|
TF:M03970
|
Factor: ELK1; motif: ACCGGAAGTN
|
0.0176895448799723
|
4782
|
69
|
29
|
0.420289855072464
|
0.00606440819740694
|
|
TF
|
TF:M04183
|
Factor: Ngn-2; motif: RACATATGTC
|
0.0181570916196269
|
1070
|
9
|
4
|
0.444444444444444
|
0.00373831775700935
|
|
TF
|
TF:M04436_1
|
Factor: RHOXF1; motif: GGMTNATCC; match class: 1
|
0.0181641064201969
|
397
|
10
|
3
|
0.3
|
0.00755667506297229
|
|
TF
|
TF:M10086_1
|
Factor: TAFII250; motif: RARRWGGCGGMGGNGR; match class: 1
|
0.0183713587106123
|
4153
|
141
|
46
|
0.326241134751773
|
0.0110763303635926
|
|
TF
|
TF:M01876
|
Factor: GABP-beta; motif: ASMGGAAGKGN
|
0.0184173911668993
|
2138
|
69
|
17
|
0.246376811594203
|
0.00795135640785781
|
|
TF
|
TF:M03916
|
Factor: PRDM1; motif: NRAAAGTGAAAGTNN
|
0.0185578501608904
|
948
|
124
|
15
|
0.120967741935484
|
0.0158227848101266
|
|
TF
|
TF:M10535
|
Factor: znf136; motif: CAAGAATWCTATAYCCAG
|
0.0186799883366663
|
8301
|
78
|
47
|
0.602564102564103
|
0.00566196843753765
|
|
TF
|
TF:M00426
|
Factor: E2F; motif: TTTSGCGS
|
0.0190075556289597
|
11885
|
114
|
85
|
0.745614035087719
|
0.00715187210769878
|
|
TF
|
TF:M11005
|
Factor: LHX4; motif: NNCRTTAN
|
0.0190075556289597
|
10614
|
91
|
64
|
0.703296703296703
|
0.00602977199924628
|
|
TF
|
TF:M01759
|
Factor: AML2; motif: AACCRCAA
|
0.0190075556289597
|
3027
|
68
|
21
|
0.308823529411765
|
0.00693756194251734
|
|
TF
|
TF:M00416_1
|
Factor: Cart-1; motif: NNNTAATTNNCATTANCN; match class: 1
|
0.0190075556289597
|
916
|
5
|
3
|
0.6
|
0.00327510917030568
|
|
TF
|
TF:M08467
|
Factor: Erm:HOXB13; motif: RSCGGAWGTNRTWAA
|
0.0190075556289597
|
706
|
71
|
9
|
0.126760563380282
|
0.0127478753541076
|
|
TF
|
TF:M01383_1
|
Factor: Nkx3A; motif: NNNNAAGTACTTAAANN; match class: 1
|
0.0190915987661595
|
1216
|
98
|
15
|
0.153061224489796
|
0.0123355263157895
|
|
TF
|
TF:M10740_1
|
Factor: HOXB2; motif: NYAATTAN; match class: 1
|
0.0192734737416198
|
590
|
102
|
10
|
0.0980392156862745
|
0.0169491525423729
|
|
TF
|
TF:M11637
|
Factor: Sox-12; motif: ACCGAACAATN
|
0.0194544492454881
|
217
|
103
|
6
|
0.058252427184466
|
0.0276497695852535
|
|
TF
|
TF:M08473
|
Factor: FOXJ2:HOXB13; motif: TTTTATNDGNMAACA
|
0.0194544492454881
|
1653
|
45
|
11
|
0.244444444444444
|
0.00665456745311555
|
|
TF
|
TF:M04390
|
Factor: LHX9; motif: NYAATTAN
|
0.0195217286526354
|
1323
|
25
|
7
|
0.28
|
0.00529100529100529
|
|
TF
|
TF:M04439
|
Factor: SHOX2; motif: NYAATTAN
|
0.0195217286526354
|
1323
|
25
|
7
|
0.28
|
0.00529100529100529
|
|
TF
|
TF:M11307_1
|
Factor: ATF-2; motif: NNTGACGTMANN; match class: 1
|
0.0195217286526354
|
1946
|
76
|
17
|
0.223684210526316
|
0.00873586844809866
|
|
TF
|
TF:M04435
|
Factor: RAX; motif: NNYAATTANN
|
0.0195217286526354
|
1276
|
8
|
4
|
0.5
|
0.00313479623824451
|
|
TF
|
TF:M03958
|
Factor: E2F2; motif: AAAAATGGCGCCAAAAWG
|
0.0195720857583824
|
10850
|
66
|
49
|
0.742424242424242
|
0.00451612903225806
|
|
TF
|
TF:M01325
|
Factor: LHX2; motif: WNNACTAATTAGNNNNN
|
0.0196404621833171
|
1492
|
124
|
20
|
0.161290322580645
|
0.0134048257372654
|
|
TF
|
TF:M00999_1
|
Factor: AIRE; motif: WTNNNWNNTGGWWNNNWNGGNNWNWN; match class: 1
|
0.019698127674306
|
392
|
23
|
4
|
0.173913043478261
|
0.0102040816326531
|
|
TF
|
TF:M10569_1
|
Factor: HB9; motif: NTCRTTAN; match class: 1
|
0.0198477195754995
|
419
|
10
|
3
|
0.3
|
0.00715990453460621
|
|
TF
|
TF:M08822
|
Factor: Msx-1; motif: WNGNAATTANV
|
0.0198477195754995
|
6821
|
36
|
22
|
0.611111111111111
|
0.00322533352880809
|
|
TF
|
TF:M10770
|
Factor: HOXB9; motif: GYMATAAAAN
|
0.0200259771158259
|
989
|
10
|
4
|
0.4
|
0.00404448938321537
|
|
TF
|
TF:M10941_1
|
Factor: VSX1; motif: CYAATTRN; match class: 1
|
0.0202531802457061
|
799
|
64
|
9
|
0.140625
|
0.0112640801001252
|
|
TF
|
TF:M04111
|
Factor: RUNX3; motif: WAACCRCAAA
|
0.0203659708725198
|
485
|
65
|
7
|
0.107692307692308
|
0.0144329896907217
|
|
TF
|
TF:M10754
|
Factor: hoxd1; motif: NTAATTAS
|
0.0204489106128191
|
1980
|
102
|
21
|
0.205882352941176
|
0.0106060606060606
|
|
TF
|
TF:M10753
|
Factor: HOXA1; motif: NTAATTAN
|
0.0204489106128191
|
1980
|
102
|
21
|
0.205882352941176
|
0.0106060606060606
|
|
TF
|
TF:M04340
|
Factor: HOXA10; motif: NGYMATAAAAN
|
0.0204489106128191
|
1978
|
33
|
10
|
0.303030303030303
|
0.00505561172901921
|
|
TF
|
TF:M00456
|
Factor: FAC1; motif: NNNCAMAACACRNA
|
0.0204489106128191
|
8019
|
91
|
52
|
0.571428571428571
|
0.00648459907719167
|
|
TF
|
TF:M08771
|
Factor: HOXA4; motif: RYCATTRN
|
0.0211786607743334
|
2900
|
13
|
7
|
0.538461538461538
|
0.00241379310344828
|
|
TF
|
TF:M09659
|
Factor: PU.1; motif: NRAAAGAGGAAGTGARA
|
0.021399298851304
|
2523
|
11
|
6
|
0.545454545454545
|
0.00237812128418549
|
|
TF
|
TF:M10694_1
|
Factor: HOXA6; motif: NGYMATTANN; match class: 1
|
0.0214195621025338
|
702
|
34
|
6
|
0.176470588235294
|
0.00854700854700855
|
|
TF
|
TF:M11225
|
Factor: CREM; motif: NNTGACGTCANN
|
0.0214195621025338
|
2710
|
140
|
33
|
0.235714285714286
|
0.0121771217712177
|
|
TF
|
TF:M11133
|
Factor: Ngn-2; motif: RMCATATGYY
|
0.0214195621025338
|
1487
|
66
|
13
|
0.196969696969697
|
0.00874243443174176
|
|
TF
|
TF:M10609_1
|
Factor: Dlx-3; motif: NTCRTTAN; match class: 1
|
0.0214442574311926
|
913
|
11
|
4
|
0.363636363636364
|
0.00438116100766703
|
|
TF
|
TF:M10698_1
|
Factor: HOXB5; motif: RTCRTTAN; match class: 1
|
0.0214442574311926
|
913
|
11
|
4
|
0.363636363636364
|
0.00438116100766703
|
|
TF
|
TF:M01863
|
Factor: ATF-3; motif: RTKRCGTCANN
|
0.0214442574311926
|
1885
|
122
|
23
|
0.188524590163934
|
0.0122015915119363
|
|
TF
|
TF:M11151
|
Factor: MSGN1; motif: NRCCAWWTGKYN
|
0.0214442574311926
|
353
|
66
|
6
|
0.0909090909090909
|
0.0169971671388102
|
|
TF
|
TF:M00731
|
Factor: Osf2; motif: ACCACANM
|
0.0214442574311926
|
1303
|
66
|
12
|
0.181818181818182
|
0.00920951650038373
|
|
TF
|
TF:M00430_1
|
Factor: E2F-1; motif: NTTSGCGG; match class: 1
|
0.0216910858436541
|
2524
|
71
|
19
|
0.267605633802817
|
0.00752773375594295
|
|
TF
|
TF:M11041
|
Factor: Pbx1; motif: KTGATKGAYN
|
0.0217370056088713
|
2087
|
3
|
3
|
1
|
0.00143747005270724
|
|
TF
|
TF:M08947
|
Factor: C-FOS; motif: NNNKATGACGTCATNNN
|
0.0220085557285322
|
7042
|
77
|
41
|
0.532467532467532
|
0.00582220959954558
|
|
TF
|
TF:M00939
|
Factor: E2F; motif: TTTSGCGSG
|
0.0220085557285322
|
6433
|
134
|
61
|
0.455223880597015
|
0.0094823565987875
|
|
TF
|
TF:M04110_1
|
Factor: RUNX3; motif: NRACCGCANWAACCRCAN; match class: 1
|
0.022159393621469
|
3764
|
140
|
42
|
0.3
|
0.0111583421891605
|
|
TF
|
TF:M10766
|
Factor: Meox2; motif: NYMATTAN
|
0.0223773424612816
|
852
|
74
|
10
|
0.135135135135135
|
0.0117370892018779
|
|
TF
|
TF:M10570
|
Factor: HB9; motif: NYAATTAN
|
0.0223773424612816
|
852
|
74
|
10
|
0.135135135135135
|
0.0117370892018779
|
|
TF
|
TF:M10732
|
Factor: HOXA2; motif: NTAATTAN
|
0.0223773424612816
|
852
|
74
|
10
|
0.135135135135135
|
0.0117370892018779
|
|
TF
|
TF:M11006
|
Factor: LHX4; motif: NTAATTAS
|
0.0223773424612816
|
852
|
74
|
10
|
0.135135135135135
|
0.0117370892018779
|
|
TF
|
TF:M10734
|
Factor: HOXA2; motif: NTAATTAN
|
0.0223773424612816
|
852
|
74
|
10
|
0.135135135135135
|
0.0117370892018779
|
|
TF
|
TF:M11414
|
Factor: GABP-alpha; motif: NACCGGAAGTN
|
0.0223773424612816
|
2907
|
49
|
16
|
0.326530612244898
|
0.00550395596835225
|
|
TF
|
TF:M10762
|
Factor: MOX1; motif: NYAATTAN
|
0.0223773424612816
|
852
|
74
|
10
|
0.135135135135135
|
0.0117370892018779
|
|
TF
|
TF:M04348
|
Factor: HOXB3; motif: NNYMATTANN
|
0.0224977704596049
|
1393
|
71
|
13
|
0.183098591549296
|
0.00933237616654702
|
|
TF
|
TF:M12334
|
Factor: ZBTB20; motif: NNAAYGTATRKN
|
0.0225037726668733
|
1214
|
62
|
11
|
0.17741935483871
|
0.00906095551894563
|
|
TF
|
TF:M11999
|
Factor: AML3; motif: NWAACCACRAAAACCACRAN
|
0.0225037726668733
|
6894
|
34
|
21
|
0.617647058823529
|
0.0030461270670148
|
|
TF
|
TF:M00916
|
Factor: CREB; motif: NNTKACGTCANNNS
|
0.0225037726668733
|
5889
|
77
|
36
|
0.467532467532468
|
0.00611309220580744
|
|
TF
|
TF:M00460_1
|
Factor: STAT5A; motif: TTCCNRGAANNNNNNTTCCNNGRR; match class: 1
|
0.0225037726668733
|
2567
|
5
|
4
|
0.8
|
0.00155823918971562
|
|
TF
|
TF:M00981_1
|
Factor: CREB,; motif: NTGACGTNA; match class: 1
|
0.0225469922337844
|
2904
|
77
|
22
|
0.285714285714286
|
0.00757575757575758
|
|
TF
|
TF:M11526
|
Factor: E2F-3; motif: NTTTTGGCGCCAAAAN
|
0.0226136753863048
|
7178
|
33
|
21
|
0.636363636363636
|
0.00292560601838952
|
|
TF
|
TF:M10727_1
|
Factor: HOXB8; motif: RTCRTTAN; match class: 1
|
0.0226136753863048
|
1379
|
13
|
5
|
0.384615384615385
|
0.00362581580855693
|
|
TF
|
TF:M10780
|
Factor: HOXC9; motif: GYMATAAAAN
|
0.0226136753863048
|
458
|
10
|
3
|
0.3
|
0.00655021834061135
|
|
TF
|
TF:M04443
|
Factor: UNCX; motif: NYAATTAN
|
0.0226136753863048
|
3248
|
119
|
33
|
0.277310924369748
|
0.0101600985221675
|
|
TF
|
TF:M10778
|
Factor: hoxd9; motif: GYMATAAAAN
|
0.0226136753863048
|
458
|
10
|
3
|
0.3
|
0.00655021834061135
|
|
TF
|
TF:M10593_1
|
Factor: Dlx-6; motif: NTCRTTAN; match class: 1
|
0.0226136753863048
|
1791
|
77
|
16
|
0.207792207792208
|
0.00893355667225014
|
|
TF
|
TF:M10600_1
|
Factor: Dlx-7; motif: NTCRTTAN; match class: 1
|
0.0226136753863048
|
2110
|
13
|
6
|
0.461538461538462
|
0.0028436018957346
|
|
TF
|
TF:M10773
|
Factor: hoxd9; motif: GYMATAAAAN
|
0.0226136753863048
|
458
|
10
|
3
|
0.3
|
0.00655021834061135
|
|
TF
|
TF:M07314
|
Factor: Blimp-1; motif: RGRAAGNGAAAG
|
0.0233609224751303
|
1460
|
94
|
16
|
0.170212765957447
|
0.010958904109589
|
|
TF
|
TF:M11220_1
|
Factor: CREB1; motif: NNTGACGTCANN; match class: 1
|
0.0233609224751303
|
1713
|
52
|
12
|
0.230769230769231
|
0.00700525394045534
|
|
TF
|
TF:M09612
|
Factor: EVX2; motif: TTTTATKRCNNN
|
0.0233609224751303
|
1070
|
10
|
4
|
0.4
|
0.00373831775700935
|
|
TF
|
TF:M04744_1
|
Factor: ATF-3; motif: GGCGCSSNSNGRTSACGTSA; match class: 1
|
0.0233609224751303
|
4889
|
140
|
51
|
0.364285714285714
|
0.0104315811004295
|
|
TF
|
TF:M09653
|
Factor: AML1; motif: NYTGTGGTTWN
|
0.0233609224751303
|
1352
|
65
|
12
|
0.184615384615385
|
0.00887573964497041
|
|
TF
|
TF:M12057
|
Factor: ZBP99; motif: GCCCMTCCCCCR
|
0.0233609224751303
|
5225
|
141
|
54
|
0.382978723404255
|
0.0103349282296651
|
|
TF
|
TF:M04347
|
Factor: HOXB2; motif: NNTMATTANN
|
0.023496540738289
|
3395
|
119
|
34
|
0.285714285714286
|
0.0100147275405007
|
|
TF
|
TF:M00017
|
Factor: ATF; motif: CNSTGACGTNNNYC
|
0.023496540738289
|
7226
|
78
|
42
|
0.538461538461538
|
0.00581234431220592
|
|
TF
|
TF:M09976
|
Factor: LHX2; motif: NNYTAATTRNNN
|
0.0235815145342972
|
2364
|
124
|
27
|
0.217741935483871
|
0.0114213197969543
|
|
TF
|
TF:M07250
|
Factor: E2F-1; motif: NNNSSCGCSAANN
|
0.0237934048553962
|
10703
|
142
|
95
|
0.669014084507042
|
0.00887601607026067
|
|
TF
|
TF:M10611
|
Factor: Dlx-5; motif: NYAATTAN
|
0.0239402758493974
|
473
|
10
|
3
|
0.3
|
0.00634249471458774
|
|
TF
|
TF:M11220
|
Factor: CREB1; motif: NNTGACGTCANN
|
0.0240757912911826
|
1725
|
52
|
12
|
0.230769230769231
|
0.00695652173913044
|
|
TF
|
TF:M10691
|
Factor: HOXB6; motif: NTAATKRC
|
0.024309977555778
|
5329
|
76
|
33
|
0.434210526315789
|
0.00619253143178833
|
|
TF
|
TF:M08222_1
|
Factor: Elk-1:HOXB13; motif: ACCGGAAGTNGTAAAN; match class: 1
|
0.0243496664255412
|
2063
|
81
|
18
|
0.222222222222222
|
0.00872515753756665
|
|
TF
|
TF:M03797
|
Factor: Msx-2; motif: TWWTTGGDGABN
|
0.0243496664255412
|
4889
|
124
|
46
|
0.370967741935484
|
0.00940887707097566
|
|
TF
|
TF:M12341
|
Factor: BCL-6; motif: NYGCTTTCKAGGAANN
|
0.0243496664255412
|
6051
|
68
|
33
|
0.485294117647059
|
0.00545364402578086
|
|
TF
|
TF:M02279
|
Factor: CREB1; motif: TGACGTCA
|
0.0243496664255412
|
1731
|
52
|
12
|
0.230769230769231
|
0.00693240901213172
|
|
TF
|
TF:M10716
|
Factor: HOXA4; motif: RTMATTAN
|
0.0243496664255412
|
4050
|
13
|
8
|
0.615384615384615
|
0.00197530864197531
|
|
TF
|
TF:M10873
|
Factor: alx3; motif: NYAATTAN
|
0.0245965394783229
|
3529
|
124
|
36
|
0.290322580645161
|
0.0102011901388495
|
|
TF
|
TF:M11582
|
Factor: FOXI1; motif: AAYRTAAACAAT
|
0.0245965394783229
|
1155
|
130
|
17
|
0.130769230769231
|
0.0147186147186147
|
|
TF
|
TF:M04112
|
Factor: GMEB2; motif: TTACGYAM
|
0.0245965394783229
|
292
|
84
|
6
|
0.0714285714285714
|
0.0205479452054795
|
|
TF
|
TF:M02084_1
|
Factor: AML1; motif: TGTGGT; match class: 1
|
0.0246727819872213
|
5429
|
5
|
5
|
1
|
0.000920979922637686
|
|
TF
|
TF:M00271_1
|
Factor: AML1a; motif: TGTGGT; match class: 1
|
0.0246727819872213
|
5429
|
5
|
5
|
1
|
0.000920979922637686
|
|
TF
|
TF:M11039
|
Factor: Pbx1; motif: NTGATTGAYR
|
0.0246727819872213
|
2796
|
14
|
7
|
0.5
|
0.0025035765379113
|
|
TF
|
TF:M11381_1
|
Factor: ESE-1; motif: SATKGCGGATGCN; match class: 1
|
0.0246727819872213
|
9745
|
99
|
64
|
0.646464646464647
|
0.00656747049769112
|
|
TF
|
TF:M00233
|
Factor: MEF-2A; motif: NNTGTTACTAAAAATAGAAMNN
|
0.0246727819872213
|
1097
|
5
|
3
|
0.6
|
0.00273473108477666
|
|
TF
|
TF:M00751_1
|
Factor: AML1; motif: TGTGGT; match class: 1
|
0.0246727819872213
|
5429
|
5
|
5
|
1
|
0.000920979922637686
|
|
TF
|
TF:M01452
|
Factor: HOXA5; motif: ANGNTAATTANCNNAN
|
0.0246727819872213
|
3709
|
74
|
25
|
0.337837837837838
|
0.00674036128336479
|
|
TF
|
TF:M09600
|
Factor: CREB1; motif: TKACGTCAYNN
|
0.0247175611792555
|
5324
|
27
|
15
|
0.555555555555556
|
0.00281743050338092
|
|
TF
|
TF:M11976_1
|
Factor: RFX7; motif: CGTTGCYAY; match class: 1
|
0.0249170254365302
|
315
|
32
|
4
|
0.125
|
0.0126984126984127
|
|
TF
|
TF:M07370_1
|
Factor: YY1; motif: NNNNAARATGGNNNN; match class: 1
|
0.0249170254365302
|
1076
|
116
|
15
|
0.129310344827586
|
0.0139405204460967
|
|
TF
|
TF:M10941
|
Factor: VSX1; motif: CYAATTRN
|
0.0249170254365302
|
3970
|
127
|
40
|
0.31496062992126
|
0.0100755667506297
|
|
TF
|
TF:M02281_1
|
Factor: SP1; motif: CCCCKCCCCC; match class: 1
|
0.0249170254365302
|
2558
|
116
|
27
|
0.232758620689655
|
0.0105551211884285
|
|
TF
|
TF:M01412_1
|
Factor: Msx-1; motif: NNNNANTAATTANTNN; match class: 1
|
0.0251150654772717
|
1264
|
9
|
4
|
0.444444444444444
|
0.00316455696202532
|
|
TF
|
TF:M10858_1
|
Factor: En-2; motif: NTCRTTARN; match class: 1
|
0.0255608575149888
|
3267
|
66
|
21
|
0.318181818181818
|
0.00642791551882461
|
|
TF
|
TF:M02266
|
Factor: HNF1B; motif: TTAATRWTTAAC
|
0.0258856656844038
|
499
|
10
|
3
|
0.3
|
0.00601202404809619
|
|
TF
|
TF:M08774
|
Factor: HOXD4; motif: NNYMATTANN
|
0.0258856656844038
|
4001
|
4
|
4
|
1
|
0.000999750062484379
|
|
TF
|
TF:M01438_1
|
Factor: ipf1; motif: NNRNTAATTAGYNCAN; match class: 1
|
0.0259142329381402
|
1067
|
62
|
10
|
0.161290322580645
|
0.00937207122774133
|
|
TF
|
TF:M01011_1
|
Factor: HNF-1; motif: NNNNGNTAAWNATTAACYNNN; match class: 1
|
0.0259142329381402
|
342
|
30
|
4
|
0.133333333333333
|
0.0116959064327485
|
|
TF
|
TF:M11932_1
|
Factor: Oct3; motif: NNTTATGYWAATKARN; match class: 1
|
0.0259142329381402
|
1127
|
5
|
3
|
0.6
|
0.00266193433895297
|
|
TF
|
TF:M03974
|
Factor: ERF; motif: ACCGGAAGTR
|
0.0260843275921997
|
956
|
69
|
10
|
0.144927536231884
|
0.0104602510460251
|
|
TF
|
TF:M08338
|
Factor: HOXA3:TBR2; motif: TAATGAGGTGTNA
|
0.0260843275921997
|
1151
|
31
|
7
|
0.225806451612903
|
0.00608166811468288
|
|
TF
|
TF:M08947_1
|
Factor: C-FOS; motif: NNNKATGACGTCATNNN; match class: 1
|
0.0261560198564443
|
2723
|
44
|
14
|
0.318181818181818
|
0.0051413881748072
|
|
TF
|
TF:M08004
|
Factor: SHOX; motif: NTAATTRN
|
0.0261560198564443
|
2226
|
13
|
6
|
0.461538461538462
|
0.00269541778975741
|
|
TF
|
TF:M00980
|
Factor: TBP; motif: TTTATAN
|
0.0262305842748917
|
2781
|
5
|
4
|
0.8
|
0.00143833153541891
|
|
TF
|
TF:M01452_1
|
Factor: HOXA5; motif: ANGNTAATTANCNNAN; match class: 1
|
0.0262943519547128
|
1429
|
72
|
13
|
0.180555555555556
|
0.00909727081875437
|
|
TF
|
TF:M10800_1
|
Factor: HOXA10; motif: NGYMATAAANN; match class: 1
|
0.0262943519547128
|
509
|
10
|
3
|
0.3
|
0.00589390962671906
|
|
TF
|
TF:M10832
|
Factor: HOXA13; motif: NCCAATAAAAN
|
0.0262943519547128
|
679
|
16
|
4
|
0.25
|
0.00589101620029455
|
|
TF
|
TF:M09869
|
Factor: ATF-4; motif: RNMTGATGCAAY
|
0.0262943519547128
|
2126
|
18
|
7
|
0.388888888888889
|
0.00329256820319849
|
|
TF
|
TF:M11023_1
|
Factor: IRX2a; motif: NCRTGTWNWACAYGN; match class: 1
|
0.0262943519547128
|
1313
|
78
|
13
|
0.166666666666667
|
0.0099009900990099
|
|
TF
|
TF:M10670
|
Factor: Emx-1; motif: NTAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M10674
|
Factor: Emx-2; motif: CTAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M04316
|
Factor: EN1; motif: NYAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M10967
|
Factor: ESX1L; motif: NYAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M11585
|
Factor: FOXJ2; motif: NNTGTTGTAAAYAN
|
0.0262943519547128
|
5564
|
5
|
5
|
1
|
0.00089863407620417
|
|
TF
|
TF:M10677
|
Factor: Vax-2; motif: NTAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M10675
|
Factor: Vax-1; motif: NYMATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M00695_1
|
Factor: ETF; motif: GVGGMGG; match class: 1
|
0.0262943519547128
|
7032
|
142
|
68
|
0.47887323943662
|
0.00967007963594994
|
|
TF
|
TF:M10678
|
Factor: Vax-2; motif: NTAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M10865
|
Factor: EVX1; motif: NTAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M10866
|
Factor: EVX2; motif: NTAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M10746
|
Factor: ipf1; motif: CTAATKRC
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M04425
|
Factor: PDX1; motif: NYAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M02097
|
Factor: LHX3; motif: ATTAAW
|
0.0262943519547128
|
11840
|
92
|
69
|
0.75
|
0.0058277027027027
|
|
TF
|
TF:M10568
|
Factor: HB9; motif: NYAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M10863
|
Factor: EVX1; motif: NTAATTAN
|
0.0262943519547128
|
826
|
67
|
9
|
0.134328358208955
|
0.0108958837772397
|
|
TF
|
TF:M10739
|
Factor: HOXB2; motif: RTCATTAN
|
0.0267013831683597
|
3917
|
11
|
7
|
0.636363636363636
|
0.0017870819504723
|
|
TF
|
TF:M00041
|
Factor: ATF2:c-Jun; motif: TGACGTYA
|
0.0267013831683597
|
4294
|
52
|
21
|
0.403846153846154
|
0.00489054494643689
|
|
TF
|
TF:M10695_1
|
Factor: HOXA6; motif: RTCRTTAN; match class: 1
|
0.0269006698930265
|
1493
|
13
|
5
|
0.384615384615385
|
0.00334896182183523
|
|
TF
|
TF:M12345
|
Factor: Zbtb37; motif: NYACCGCRNTCACCGCR
|
0.0269006698930265
|
5505
|
142
|
56
|
0.394366197183099
|
0.010172570390554
|
|
TF
|
TF:M00137
|
Factor: Oct-1; motif: NNNRTAATNANNN
|
0.0269006698930265
|
6851
|
11
|
9
|
0.818181818181818
|
0.00131367683549847
|
|
TF
|
TF:M09839
|
Factor: ATF4; motif: ATTGCATCAT
|
0.0269006698930265
|
2145
|
18
|
7
|
0.388888888888889
|
0.00326340326340326
|
|
TF
|
TF:M10638
|
Factor: Nkx2-3; motif: NNCGTTRWS
|
0.0269006698930265
|
10804
|
109
|
75
|
0.688073394495413
|
0.00694187338022954
|
|
TF
|
TF:M07483
|
Factor: CXXC1; motif: KNRNAMCGMWAAMNN
|
0.0269006698930265
|
1318
|
9
|
4
|
0.444444444444444
|
0.00303490136570561
|
|
TF
|
TF:M09777_1
|
Factor: NR1H4; motif: NNAATGACCNN; match class: 1
|
0.0269418028881034
|
522
|
10
|
3
|
0.3
|
0.00574712643678161
|
|
TF
|
TF:M01856
|
Factor: AML3; motif: AACCACAN
|
0.0272373002543252
|
1198
|
66
|
11
|
0.166666666666667
|
0.00918196994991653
|
|
TF
|
TF:M04298
|
Factor: Cdx-2; motif: GYMATAAAA
|
0.027332052306413
|
1174
|
10
|
4
|
0.4
|
0.00340715502555366
|
|
TF
|
TF:M01199_1
|
Factor: RNF96; motif: BCCCGCRGCC; match class: 1
|
0.0274752037319274
|
4322
|
104
|
36
|
0.346153846153846
|
0.00832947709393799
|
|
TF
|
TF:M11152
|
Factor: MSGN1; motif: NRCCAWWTGKYN
|
0.0277050777851115
|
534
|
66
|
7
|
0.106060606060606
|
0.0131086142322097
|
|
TF
|
TF:M10802_1
|
Factor: HOXA10; motif: NGYMATAAANN; match class: 1
|
0.0277050777851115
|
530
|
10
|
3
|
0.3
|
0.00566037735849057
|
|
TF
|
TF:M10742
|
Factor: Hox-D3; motif: GTCRTTAR
|
0.0277050777851115
|
530
|
10
|
3
|
0.3
|
0.00566037735849057
|
|
TF
|
TF:M10709
|
Factor: HOXD4; motif: NYMATKAN
|
0.0277050777851115
|
530
|
10
|
3
|
0.3
|
0.00566037735849057
|
|
TF
|
TF:M11207_1
|
Factor: AP-4; motif: ANCATATGNT; match class: 1
|
0.0281847774813031
|
2394
|
30
|
10
|
0.333333333333333
|
0.00417710944026733
|
|
TF
|
TF:M04012
|
Factor: HSFY1; motif: TTTCGAACG
|
0.0286644718880533
|
1013
|
139
|
16
|
0.115107913669065
|
0.0157946692991115
|
|
TF
|
TF:M00338
|
Factor: ATF; motif: NTGACGTCANYS
|
0.0291215836132953
|
3798
|
70
|
24
|
0.342857142857143
|
0.00631911532385466
|
|
TF
|
TF:M03929
|
Factor: ZBTB7C; motif: NCGACCACCGNN
|
0.0291215836132953
|
2959
|
29
|
11
|
0.379310344827586
|
0.00371747211895911
|
|
TF
|
TF:M10657
|
Factor: NKX6B; motif: NTMATTAW
|
0.0291215836132953
|
2650
|
46
|
14
|
0.304347826086957
|
0.00528301886792453
|
|
TF
|
TF:M10578
|
Factor: Barhl-1; motif: NTAAACGN
|
0.0291215836132953
|
6099
|
61
|
30
|
0.491803278688525
|
0.00491883915395967
|
|
TF
|
TF:M04321_1
|
Factor: EVX2; motif: NNTNATTANN; match class: 1
|
0.0291215836132953
|
1477
|
71
|
13
|
0.183098591549296
|
0.00880162491536899
|
|
TF
|
TF:M01400
|
Factor: Dlx-3; motif: NNNNNATAATTACMNNN
|
0.0299257993675605
|
3276
|
23
|
10
|
0.434782608695652
|
0.00305250305250305
|
|
TF
|
TF:M03873
|
Factor: HOXA10; motif: ANTTTATG
|
0.0299331261302914
|
2291
|
100
|
22
|
0.22
|
0.00960279353993889
|
|
TF
|
TF:M02026_1
|
Factor: MEF-2D; motif: WAAATAR; match class: 1
|
0.0299445893163857
|
2408
|
9
|
5
|
0.555555555555556
|
0.00207641196013289
|
|
TF
|
TF:M10799_1
|
Factor: HOXA10; motif: NRTCGTAAANN; match class: 1
|
0.0301271127462592
|
1078
|
108
|
14
|
0.12962962962963
|
0.012987012987013
|
|
TF
|
TF:M01861
|
Factor: ATF-1; motif: TNACGTCAN
|
0.0301271127462592
|
6294
|
77
|
37
|
0.480519480519481
|
0.00587861455354306
|
|
TF
|
TF:M00161
|
Factor: Oct-1; motif: MKVATTTGCATATT
|
0.0304646310574177
|
2978
|
5
|
4
|
0.8
|
0.00134318334452653
|
|
TF
|
TF:M09946
|
Factor: HNF-1beta; motif: NGTTAATRATTAACN
|
0.0309446948430215
|
833
|
33
|
6
|
0.181818181818182
|
0.00720288115246098
|
|
TF
|
TF:M11987
|
Factor: NFATc3; motif: NAYGGAAANW
|
0.0310465235696164
|
2409
|
132
|
28
|
0.212121212121212
|
0.0116230801162308
|
|
TF
|
TF:M08894
|
Factor: OVOL; motif: ANRTAACGG
|
0.0310465235696164
|
8109
|
72
|
42
|
0.583333333333333
|
0.00517943026267111
|
|
TF
|
TF:M10681
|
Factor: HPX42B; motif: NNYNATTANN
|
0.0310465235696164
|
2469
|
117
|
26
|
0.222222222222222
|
0.010530579181855
|
|
TF
|
TF:M01019
|
Factor: Tbx5; motif: NNAGGTGTNANN
|
0.0310465235696164
|
5880
|
5
|
5
|
1
|
0.000850340136054422
|
|
TF
|
TF:M08203
|
Factor: CDP:T-bet; motif: NTCACACNYYRATCRATM
|
0.031719519802139
|
6147
|
34
|
19
|
0.558823529411765
|
0.00309093866926956
|
|
TF
|
TF:M00739_1
|
Factor: E2F-4:DP-2; motif: TTTCSCGC; match class: 1
|
0.0318830514522182
|
1187
|
132
|
17
|
0.128787878787879
|
0.0143218197135636
|
|
TF
|
TF:M08470
|
Factor: Fli-1:Dlx-2; motif: RNCGGAARYAATTA
|
0.0321789340946574
|
5930
|
5
|
5
|
1
|
0.000843170320404722
|
|
TF
|
TF:M08215_1
|
Factor: Elk-1:EVX1; motif: NNCGGWAATNNYMATTA; match class: 1
|
0.0323715162144952
|
1638
|
30
|
8
|
0.266666666666667
|
0.00488400488400488
|
|
TF
|
TF:M11017
|
Factor: DPRX; motif: AGATAATCCN
|
0.0324450048221687
|
930
|
13
|
4
|
0.307692307692308
|
0.0043010752688172
|
|
TF
|
TF:M08826_1
|
Factor: NF-E4; motif: GTGAGGS; match class: 1
|
0.0324947289046877
|
4695
|
49
|
21
|
0.428571428571429
|
0.00447284345047923
|
|
TF
|
TF:M10873_1
|
Factor: alx3; motif: NYAATTAN; match class: 1
|
0.0325175220654908
|
735
|
64
|
8
|
0.125
|
0.0108843537414966
|
|
TF
|
TF:M11884
|
Factor: Pax-4a; motif: TTYACGCWTSANYGMACN
|
0.0325375772773479
|
348
|
133
|
8
|
0.0601503759398496
|
0.0229885057471264
|
|
TF
|
TF:M04402
|
Factor: MIXL1; motif: NNYAATTANN
|
0.0327082701788122
|
2460
|
124
|
27
|
0.217741935483871
|
0.0109756097560976
|
|
TF
|
TF:M12005
|
Factor: GMEB-1; motif: KNACGTNRNNACGTAN
|
0.0329399641219618
|
1166
|
70
|
11
|
0.157142857142857
|
0.00943396226415094
|
|
TF
|
TF:M04194
|
Factor: TFEB; motif: RNCACGTGAC
|
0.0334441281560545
|
1436
|
139
|
20
|
0.143884892086331
|
0.0139275766016713
|
|
TF
|
TF:M10687
|
Factor: HOXB7; motif: NGTAATTANN
|
0.033493386098373
|
942
|
13
|
4
|
0.307692307692308
|
0.00424628450106157
|
|
TF
|
TF:M01400_1
|
Factor: Dlx-3; motif: NNNNNATAATTACMNNN; match class: 1
|
0.033493386098373
|
944
|
30
|
6
|
0.2
|
0.00635593220338983
|
|
TF
|
TF:M11896_1
|
Factor: Oct-2; motif: NTATGCWAATN; match class: 1
|
0.0335913874825417
|
1308
|
5
|
3
|
0.6
|
0.00229357798165138
|
|
TF
|
TF:M04307
|
Factor: DPRX; motif: NRGATAATCCN
|
0.0336832479061494
|
883
|
32
|
6
|
0.1875
|
0.00679501698754247
|
|
TF
|
TF:M01408_1
|
Factor: Brn-3c; motif: NNTTATTAATKANKNC; match class: 1
|
0.033730992147989
|
1930
|
63
|
14
|
0.222222222222222
|
0.00725388601036269
|
|
TF
|
TF:M10558
|
Factor: Prox-1; motif: NAAGGCGTCNTN
|
0.0337975483957149
|
996
|
59
|
9
|
0.152542372881356
|
0.00903614457831325
|
|
TF
|
TF:M08773_1
|
Factor: HOXA7; motif: NYMATTAN; match class: 1
|
0.033883615543524
|
830
|
34
|
6
|
0.176470588235294
|
0.0072289156626506
|
|
TF
|
TF:M10712
|
Factor: HOXD4; motif: NYMATTAN
|
0.033994101011038
|
4154
|
11
|
7
|
0.636363636363636
|
0.00168512277323062
|
|
TF
|
TF:M02013
|
Factor: HNF-1alpha; motif: RGTTAATNWTT
|
0.033994101011038
|
1522
|
47
|
10
|
0.212765957446809
|
0.00657030223390276
|
|
TF
|
TF:M10886
|
Factor: PRX-2; motif: NYAATTAN
|
0.0340422202155142
|
1156
|
25
|
6
|
0.24
|
0.00519031141868512
|
|
TF
|
TF:M11313
|
Factor: CREBPA; motif: NRTGACGTMANN
|
0.0340422202155142
|
3680
|
28
|
12
|
0.428571428571429
|
0.00326086956521739
|
|
TF
|
TF:M00457
|
Factor: STAT5A; motif: NAWTTCYNGGAANYN
|
0.0344184024423406
|
4038
|
60
|
22
|
0.366666666666667
|
0.00544824170381377
|
|
TF
|
TF:M10104_1
|
Factor: YY1; motif: CAARATGGCGGC; match class: 1
|
0.0344803684983056
|
1138
|
116
|
15
|
0.129310344827586
|
0.0131810193321617
|
|
TF
|
TF:M04378_1
|
Factor: IRX5; motif: NWACAYRACAWN; match class: 1
|
0.0344853186766287
|
592
|
10
|
3
|
0.3
|
0.00506756756756757
|
|
TF
|
TF:M09608_1
|
Factor: Erg; motif: NNACCGGAARTSN; match class: 1
|
0.0345010555643884
|
2389
|
80
|
19
|
0.2375
|
0.00795311845960653
|
|
TF
|
TF:M08318_1
|
Factor: GCMa:FOXO1A; motif: GTMAATAMGGGTRN; match class: 1
|
0.0349703970733162
|
4671
|
15
|
9
|
0.6
|
0.00192678227360308
|
|
TF
|
TF:M10924_1
|
Factor: Gscl; motif: NTAATCCN; match class: 1
|
0.0349732652721348
|
1631
|
13
|
5
|
0.384615384615385
|
0.00306560392397302
|
|
TF
|
TF:M10612_1
|
Factor: Dlx-5; motif: NTCRTTAN; match class: 1
|
0.0351656496045489
|
2544
|
25
|
9
|
0.36
|
0.0035377358490566
|
|
TF
|
TF:M11000
|
Factor: LIM-1; motif: NYAATTAN
|
0.0351656496045489
|
5830
|
31
|
17
|
0.548387096774194
|
0.00291595197255575
|
|
TF
|
TF:M10603_1
|
Factor: Dlx-7; motif: NTCRTTAN; match class: 1
|
0.0351656496045489
|
2544
|
25
|
9
|
0.36
|
0.0035377358490566
|
|
TF
|
TF:M00999
|
Factor: AIRE; motif: WTNNNWNNTGGWWNNNWNGGNNWNWN
|
0.0353222417172461
|
3389
|
13
|
7
|
0.538461538461538
|
0.002065506048982
|
|
TF
|
TF:M10822
|
Factor: HOXA11; motif: NGYMATAAAAN
|
0.0354027416146258
|
2237
|
10
|
5
|
0.5
|
0.00223513634331694
|
|
TF
|
TF:M04305
|
Factor: DMBX1; motif: NNGGATTANN
|
0.0355871252230111
|
5134
|
14
|
9
|
0.642857142857143
|
0.00175301908843007
|
|
TF
|
TF:M04519
|
Factor: E2F-4; motif: TTTGGCGCCAAA
|
0.0355871252230111
|
4861
|
48
|
21
|
0.4375
|
0.00432009874511417
|
|
TF
|
TF:M10584
|
Factor: Barhl2; motif: NTAAACGN
|
0.0355871252230111
|
1886
|
65
|
14
|
0.215384615384615
|
0.00742311770943796
|
|
TF
|
TF:M07611_1
|
Factor: p54NRB; motif: GRNNNMGGATGRMNNCGGA; match class: 1
|
0.0356386481884111
|
1645
|
13
|
5
|
0.384615384615385
|
0.00303951367781155
|
|
TF
|
TF:M08708
|
Factor: GCMa:pitx1; motif: GGATTAKNNNNNRTGCGGG
|
0.0358390103226275
|
581
|
65
|
7
|
0.107692307692308
|
0.0120481927710843
|
|
TF
|
TF:M00284_1
|
Factor: TCF11:MafG; motif: NNNNNATGACTCAGCANTTNNG; match class: 1
|
0.0358390103226275
|
631
|
60
|
7
|
0.116666666666667
|
0.0110935023771791
|
|
TF
|
TF:M04625
|
Factor: PLZF; motif: ACTKTANNTN
|
0.0358951723769064
|
6162
|
5
|
5
|
1
|
0.000811424862057773
|
|
TF
|
TF:M10881
|
Factor: Alx-4; motif: NYAATTAN
|
0.0358951723769064
|
1753
|
124
|
21
|
0.169354838709677
|
0.0119794637763833
|
|
TF
|
TF:M10892
|
Factor: PMX2A; motif: SYAATTAN
|
0.0358951723769064
|
1753
|
124
|
21
|
0.169354838709677
|
0.0119794637763833
|
|
TF
|
TF:M10907
|
Factor: SHOX; motif: NYAATTAN
|
0.0358951723769064
|
1753
|
124
|
21
|
0.169354838709677
|
0.0119794637763833
|
|
TF
|
TF:M04399_1
|
Factor: MEOX2; motif: NSTAATTANN; match class: 1
|
0.0360733071924856
|
880
|
23
|
5
|
0.217391304347826
|
0.00568181818181818
|
|
TF
|
TF:M10864_1
|
Factor: EVX1; motif: NTCATTAN; match class: 1
|
0.0363169747340939
|
1184
|
11
|
4
|
0.363636363636364
|
0.00337837837837838
|
|
TF
|
TF:M01337_1
|
Factor: HOXA3; motif: NNNNRNTAATTARY; match class: 1
|
0.0364026476606644
|
12033
|
117
|
86
|
0.735042735042735
|
0.00714701238261448
|
|
TF
|
TF:M01385_1
|
Factor: Pax-4; motif: WNNNYTAATTARYNSNN; match class: 1
|
0.0368776104752612
|
2495
|
124
|
27
|
0.217741935483871
|
0.0108216432865731
|
|
TF
|
TF:M00346
|
Factor: GATA-1; motif: NCWGATAACA
|
0.0375911737136686
|
4376
|
16
|
9
|
0.5625
|
0.00205667276051188
|
|
TF
|
TF:M11285_1
|
Factor: Fra-1; motif: GRTGACGTCATC; match class: 1
|
0.0375911737136686
|
4589
|
77
|
29
|
0.376623376623377
|
0.00631945957724995
|
|
TF
|
TF:M10956
|
Factor: isx; motif: NTCRTTAA
|
0.0376018208067638
|
9441
|
91
|
57
|
0.626373626373626
|
0.00603749602796314
|
|
TF
|
TF:M00746
|
Factor: Elf-1; motif: RNWMBAGGAART
|
0.0376018208067638
|
3365
|
76
|
23
|
0.302631578947368
|
0.00683506686478455
|
|
TF
|
TF:M01299
|
Factor: MECP2; motif: CCGGNNTTWA
|
0.0382398791859167
|
5228
|
108
|
42
|
0.388888888888889
|
0.0080336648814078
|
|
TF
|
TF:M10691_1
|
Factor: HOXB6; motif: NTAATKRC; match class: 1
|
0.0387299305828755
|
1497
|
34
|
8
|
0.235294117647059
|
0.0053440213760855
|
|
TF
|
TF:M03865
|
Factor: Blimp-1; motif: NRGRAAGKGAAAGK
|
0.0388782578813259
|
1048
|
58
|
9
|
0.155172413793103
|
0.00858778625954199
|
|
TF
|
TF:M03925_1
|
Factor: YY2; motif: NCCGCCATNTY; match class: 1
|
0.0393397455814326
|
765
|
141
|
13
|
0.0921985815602837
|
0.0169934640522876
|
|
TF
|
TF:M08928
|
Factor: FOSB:JUND; motif: NNTNACTNATN
|
0.0395937817913572
|
6164
|
67
|
32
|
0.477611940298507
|
0.00519143413367943
|
|
TF
|
TF:M09669_1
|
Factor: TCF-4; motif: NNNCTTTGAWSTN; match class: 1
|
0.0396090234768779
|
207
|
29
|
3
|
0.103448275862069
|
0.0144927536231884
|
|
TF
|
TF:M04611_1
|
Factor: FOXM1; motif: NAGASTGATTA; match class: 1
|
0.0398464806840376
|
3507
|
10
|
6
|
0.6
|
0.00171086398631309
|
|
TF
|
TF:M08874
|
Factor: E2F1; motif: NNNNNGCGSSAAAN
|
0.0401919875965952
|
7201
|
142
|
68
|
0.47887323943662
|
0.00944313289820858
|
|
TF
|
TF:M11922
|
Factor: Brn-4; motif: DNATGCRCATMW
|
0.0401919875965952
|
1874
|
40
|
10
|
0.25
|
0.00533617929562433
|
|
TF
|
TF:M11266_1
|
Factor: c-Jun; motif: NATGACGTCAYN; match class: 1
|
0.0403355399995736
|
6308
|
81
|
38
|
0.469135802469136
|
0.00602409638554217
|
|
TF
|
TF:M04385
|
Factor: LHX2; motif: NCTAATTANN
|
0.0403355399995736
|
900
|
135
|
14
|
0.103703703703704
|
0.0155555555555556
|
|
TF
|
TF:M10800
|
Factor: HOXA10; motif: NGYMATAAANN
|
0.0403355399995736
|
3521
|
10
|
6
|
0.6
|
0.00170406134620846
|
|
TF
|
TF:M00919
|
Factor: E2F; motif: NCSCGCSAAAN
|
0.0405801759118346
|
6149
|
134
|
57
|
0.425373134328358
|
0.00926979996747439
|
|
TF
|
TF:M11134
|
Factor: Ngn-2; motif: RACATATGTY
|
0.0406856872902287
|
1560
|
9
|
4
|
0.444444444444444
|
0.00256410256410256
|
|
TF
|
TF:M04314
|
Factor: EMX2; motif: NNTAATTANN
|
0.0406908630268785
|
247
|
117
|
6
|
0.0512820512820513
|
0.0242914979757085
|
|
TF
|
TF:M00228_1
|
Factor: VBP; motif: GTTACRTMAT; match class: 1
|
0.0406908630268785
|
442
|
66
|
6
|
0.0909090909090909
|
0.0135746606334842
|
|
TF
|
TF:M01241_1
|
Factor: BEN; motif: CWGCGAYA; match class: 1
|
0.0406908630268785
|
2097
|
106
|
21
|
0.19811320754717
|
0.0100143061516452
|
|
TF
|
TF:M04299_1
|
Factor: Dlx-1; motif: NNTAATTANN; match class: 1
|
0.0406908630268785
|
473
|
104
|
8
|
0.0769230769230769
|
0.0169133192389006
|
|
TF
|
TF:M12195
|
Factor: GATA-3; motif: MGATAASATCT
|
0.0412605745641331
|
3856
|
15
|
8
|
0.533333333333333
|
0.0020746887966805
|
|
TF
|
TF:M01379
|
Factor: HNF-1alpha; motif: NNKNNGTTAACTAANNN
|
0.0414051682419588
|
3368
|
5
|
4
|
0.8
|
0.00118764845605701
|
|
TF
|
TF:M11150_1
|
Factor: msc; motif: NRACAGCTGTYN; match class: 1
|
0.0414051682419588
|
331
|
38
|
4
|
0.105263157894737
|
0.0120845921450151
|
|
TF
|
TF:M07476
|
Factor: Lhx8; motif: TGATTG
|
0.0415707549906831
|
9096
|
19
|
15
|
0.789473684210526
|
0.0016490765171504
|
|
TF
|
TF:M11431_1
|
Factor: PEA3; motif: NTCGTAAATGCA; match class: 1
|
0.0415707549906831
|
4298
|
108
|
36
|
0.333333333333333
|
0.00837598883201489
|
|
TF
|
TF:M10667
|
Factor: NOTO; motif: NYMATTAN
|
0.0416726628402309
|
5109
|
127
|
47
|
0.37007874015748
|
0.00919945194754355
|
|
TF
|
TF:M04393
|
Factor: LMX1B; motif: NYAATTAN
|
0.0421512169876885
|
2301
|
67
|
16
|
0.238805970149254
|
0.00695349847892221
|
|
TF
|
TF:M04351
|
Factor: HOXC10; motif: GYMATWAAAN
|
0.0423955723761928
|
2121
|
11
|
5
|
0.454545454545455
|
0.00235737859500236
|
|
TF
|
TF:M09945
|
Factor: HNF-1alpha; motif: NGTTAATKATTAACY
|
0.0426182399799757
|
997
|
51
|
8
|
0.156862745098039
|
0.00802407221664995
|
|
TF
|
TF:M10858
|
Factor: En-2; motif: NTCRTTARN
|
0.0428804672339672
|
9165
|
34
|
24
|
0.705882352941177
|
0.00261865793780687
|
|
TF
|
TF:M01404
|
Factor: HOXD13; motif: NNNYYAATAAAANNNN
|
0.0430354623195971
|
4411
|
11
|
7
|
0.636363636363636
|
0.00158694173656767
|
|
TF
|
TF:M10802
|
Factor: HOXA10; motif: NGYMATAAANN
|
0.0430354623195971
|
3588
|
10
|
6
|
0.6
|
0.00167224080267559
|
|
TF
|
TF:M04402_1
|
Factor: MIXL1; motif: NNYAATTANN; match class: 1
|
0.0438931917310276
|
2055
|
116
|
22
|
0.189655172413793
|
0.010705596107056
|
|
TF
|
TF:M04320_1
|
Factor: EVX1; motif: NNTNATTANN; match class: 1
|
0.0446336616982783
|
1875
|
104
|
19
|
0.182692307692308
|
0.0101333333333333
|
|
TF
|
TF:M11587
|
Factor: foxj3; motif: RCGTMAACAN
|
0.0446405179145432
|
913
|
56
|
8
|
0.142857142857143
|
0.00876232201533406
|
|
TF
|
TF:M11273_1
|
Factor: JunB; motif: GATGACGTCAYC; match class: 1
|
0.0447451220927847
|
6582
|
81
|
39
|
0.481481481481481
|
0.00592525068368277
|
|
TF
|
TF:M10698
|
Factor: HOXB5; motif: RTCRTTAN
|
0.0448245107365591
|
4697
|
13
|
8
|
0.615384615384615
|
0.00170321481796892
|
|
TF
|
TF:M10609
|
Factor: Dlx-3; motif: NTCRTTAN
|
0.0448245107365591
|
4697
|
13
|
8
|
0.615384615384615
|
0.00170321481796892
|
|
TF
|
TF:M07055
|
Factor: Pitx1; motif: CACTAATCCYYN
|
0.0453000580832315
|
1137
|
133
|
16
|
0.120300751879699
|
0.0140721196130167
|
|
TF
|
TF:M04044
|
Factor: MYBL1; motif: NNAACCGTTAA
|
0.0453000580832315
|
515
|
142
|
10
|
0.0704225352112676
|
0.0194174757281553
|
|
TF
|
TF:M11935_1
|
Factor: Oct3; motif: NNWTATGYWAATKANN; match class: 1
|
0.0453000580832315
|
530
|
25
|
4
|
0.16
|
0.00754716981132075
|
|
TF
|
TF:M08784
|
Factor: Sox-17; motif: ACCGAACAAT
|
0.0453000580832315
|
799
|
17
|
4
|
0.235294117647059
|
0.00500625782227785
|
|
TF
|
TF:M01431
|
Factor: Barx-2; motif: TAANTAATTANTKNTN
|
0.0453000580832315
|
2185
|
71
|
16
|
0.225352112676056
|
0.00732265446224256
|
|
TF
|
TF:M00291
|
Factor: Freac-3; motif: NNNNNGTAAATAAACA
|
0.0455304818656829
|
5278
|
66
|
28
|
0.424242424242424
|
0.00530503978779841
|
|
TF
|
TF:M00477_1
|
Factor: FOXO3; motif: TNNTTGTTTACNTW; match class: 1
|
0.0456563586997808
|
277
|
47
|
4
|
0.0851063829787234
|
0.0144404332129964
|
|
TF
|
TF:M12042
|
Factor: TEF-1; motif: NRCATWCCN
|
0.0456563586997808
|
5904
|
22
|
13
|
0.590909090909091
|
0.00220189701897019
|
|
TF
|
TF:M03983
|
Factor: ETV5; motif: NCCGGAWGYN
|
0.0456692013832946
|
3129
|
69
|
20
|
0.289855072463768
|
0.00639181847235539
|
|
TF
|
TF:M04387_1
|
Factor: LHX6; motif: RCTAATTARN; match class: 1
|
0.0456692013832946
|
498
|
124
|
9
|
0.0725806451612903
|
0.0180722891566265
|
|
TF
|
TF:M04387
|
Factor: LHX6; motif: RCTAATTARN
|
0.0456692013832946
|
498
|
124
|
9
|
0.0725806451612903
|
0.0180722891566265
|
|
TF
|
TF:M00349_1
|
Factor: GATA-2; motif: ASAGATAANA; match class: 1
|
0.0458678426314653
|
673
|
32
|
5
|
0.15625
|
0.00742942050520059
|
|
TF
|
TF:M00427_1
|
Factor: E2F; motif: TTTSGCGS; match class: 1
|
0.0458878748600059
|
3831
|
113
|
34
|
0.300884955752212
|
0.00887496737144349
|
|
TF
|
TF:M03849_1
|
Factor: Sox-4; motif: AACAAA; match class: 1
|
0.0458932283830836
|
9090
|
65
|
41
|
0.630769230769231
|
0.00451045104510451
|
|
TF
|
TF:M04336
|
Factor: HNF1A; motif: NRTTAATNATTAACN
|
0.0458932283830836
|
943
|
33
|
6
|
0.181818181818182
|
0.0063626723223754
|
|
TF
|
TF:M06173
|
Factor: ZNF85; motif: KGGCGGAAGM
|
0.0459551482218047
|
100
|
130
|
4
|
0.0307692307692308
|
0.04
|
|
TF
|
TF:M10108_1
|
Factor: WT1; motif: RGGNGGGGGAGGRGGNGGRG; match class: 1
|
0.0459551482218047
|
2864
|
106
|
26
|
0.245283018867925
|
0.00907821229050279
|
|
TF
|
TF:M11895
|
Factor: Oct-2; motif: NTATGCGCATAN
|
0.0459551482218047
|
3995
|
34
|
14
|
0.411764705882353
|
0.00350438047559449
|
|
TF
|
TF:M01373
|
Factor: Cdx-1; motif: NNNGGTMATAAAANNN
|
0.0459551482218047
|
910
|
95
|
11
|
0.115789473684211
|
0.0120879120879121
|
|
TF
|
TF:M04517
|
Factor: E2F2; motif: AAAATGGCGCCATTTT
|
0.0459551482218047
|
9190
|
81
|
50
|
0.617283950617284
|
0.00544069640914037
|
|
TF
|
TF:M10685
|
Factor: HOXB7; motif: NGYAATTANN
|
0.0460136725484086
|
3549
|
74
|
23
|
0.310810810810811
|
0.0064806987883911
|
|
TF
|
TF:M10054
|
Factor: AML2; motif: NYTGTGGTTW
|
0.0460136725484086
|
201
|
65
|
4
|
0.0615384615384615
|
0.0199004975124378
|
|
TF
|
TF:M07034
|
Factor: ATF-1; motif: NNNTGACGTNNN
|
0.0460136725484086
|
4248
|
77
|
27
|
0.350649350649351
|
0.00635593220338983
|
|
TF
|
TF:M01300
|
Factor: HMGI-C; motif: ATATTSSSSNWWATT
|
0.0460136725484086
|
218
|
138
|
6
|
0.0434782608695652
|
0.0275229357798165
|
|
TF
|
TF:M10040
|
Factor: Blimp-1; motif: RAAAGNGAAAGTNN
|
0.0460702211798061
|
366
|
58
|
5
|
0.0862068965517241
|
0.0136612021857923
|
|
TF
|
TF:M11686
|
Factor: IRF-4; motif: NYGAAASYGAAACYN
|
0.0462459134254628
|
3391
|
95
|
27
|
0.284210526315789
|
0.00796225302270717
|
|
TF
|
TF:M07229
|
Factor: STAT3; motif: NTTCYKGGAAN
|
0.0462841023826229
|
3717
|
63
|
21
|
0.333333333333333
|
0.00564971751412429
|
|
TF
|
TF:M10031_1
|
Factor: ipf1; motif: TGATTGATK; match class: 1
|
0.0462841023826229
|
3647
|
13
|
7
|
0.538461538461538
|
0.00191938579654511
|
|
TF
|
TF:M00981
|
Factor: CREB,; motif: NTGACGTNA
|
0.0462946302579006
|
8345
|
119
|
65
|
0.546218487394958
|
0.00778909526662672
|
|
TF
|
TF:M04555
|
Factor: SRY; motif: AACAATANCATTGTT
|
0.0462946302579006
|
7098
|
40
|
23
|
0.575
|
0.00324034939419555
|
|
TF
|
TF:M10646_1
|
Factor: HMX2; motif: SCACTTANC; match class: 1
|
0.0462946302579006
|
1569
|
5
|
3
|
0.6
|
0.00191204588910134
|
|
TF
|
TF:M11026_1
|
Factor: Six-1; motif: NNGTATCRNN; match class: 1
|
0.0464266508517706
|
1341
|
17
|
5
|
0.294117647058824
|
0.00372856077554064
|
|
TF
|
TF:M10799
|
Factor: HOXA10; motif: NRTCGTAAANN
|
0.0465989844330183
|
5646
|
108
|
44
|
0.407407407407407
|
0.00779312787814382
|
|
TF
|
TF:M00017_1
|
Factor: ATF; motif: CNSTGACGTNNNYC; match class: 1
|
0.0465989844330183
|
2222
|
122
|
24
|
0.19672131147541
|
0.0108010801080108
|
|
TF
|
TF:M04079_1
|
Factor: POU3F2; motif: WTATGCWAATKA; match class: 1
|
0.0467782768463163
|
543
|
25
|
4
|
0.16
|
0.00736648250460405
|
|
TF
|
TF:M01376_1
|
Factor: S8; motif: ANNRCTAATTAGYNNNN; match class: 1
|
0.0467782768463163
|
298
|
102
|
6
|
0.0588235294117647
|
0.0201342281879195
|
|
TF
|
TF:M11269
|
Factor: JunD; motif: SATGACTCATN
|
0.0473799873676897
|
3913
|
28
|
12
|
0.428571428571429
|
0.00306670074111935
|
|
TF
|
TF:M10583
|
Factor: Barhl2; motif: NTAATTGN
|
0.0475400690889928
|
1104
|
38
|
7
|
0.184210526315789
|
0.00634057971014493
|
|
TF
|
TF:M02281
|
Factor: SP1; motif: CCCCKCCCCC
|
0.047586426887329
|
6480
|
116
|
52
|
0.448275862068966
|
0.00802469135802469
|
|
TF
|
TF:M11541_1
|
Factor: Foxn2; motif: NNGCGTCNNNNNGACGCNN; match class: 1
|
0.0478942957113573
|
5493
|
120
|
47
|
0.391666666666667
|
0.00855634443837612
|
|
TF
|
TF:M11325_1
|
Factor: C/EBPgamma; motif: NNTTGCGYAANN; match class: 1
|
0.0478942957113573
|
324
|
21
|
3
|
0.142857142857143
|
0.00925925925925926
|
|
TF
|
TF:M11895_1
|
Factor: Oct-2; motif: NTATGCGCATAN; match class: 1
|
0.0480363403653298
|
1376
|
31
|
7
|
0.225806451612903
|
0.00508720930232558
|
|
TF
|
TF:M10880_1
|
Factor: Alx-4; motif: NNCRTTAN; match class: 1
|
0.0481458227010339
|
2683
|
17
|
7
|
0.411764705882353
|
0.00260901975400671
|
|
TF
|
TF:M01230_1
|
Factor: ZNF333; motif: ATAAT; match class: 1
|
0.0481458227010339
|
11582
|
76
|
56
|
0.736842105263158
|
0.00483508893109998
|
|
TF
|
TF:M00004_1
|
Factor: c-Myb; motif: NCNRNNGRCNGTTGGKGG; match class: 1
|
0.0481458227010339
|
1883
|
48
|
11
|
0.229166666666667
|
0.00584174190122146
|
|
TF
|
TF:M11189
|
Factor: Myogenin; motif: YGACAGCTGTTN
|
0.0481458227010339
|
177
|
38
|
3
|
0.0789473684210526
|
0.0169491525423729
|
|
TF
|
TF:M09953
|
Factor: HOXB13; motif: TTTTATNGSN
|
0.0485694064472239
|
1570
|
21
|
6
|
0.285714285714286
|
0.00382165605095541
|
|
TF
|
TF:M04287
|
Factor: BARHL2; motif: NNTAAAYGNN
|
0.0488090050142807
|
4416
|
61
|
23
|
0.377049180327869
|
0.00520833333333333
|
|
TF
|
TF:M01837_1
|
Factor: FKLF; motif: BGGGNGGVMD; match class: 1
|
0.0488090050142807
|
1413
|
139
|
19
|
0.136690647482014
|
0.013446567586695
|
|
TF
|
TF:M00737_1
|
Factor: E2F-1:DP-2; motif: TTTSSCGC; match class: 1
|
0.0488090050142807
|
2933
|
136
|
32
|
0.235294117647059
|
0.0109103307193999
|
|
TF
|
TF:M00933_1
|
Factor: Sp1; motif: CCCCGCCCCN; match class: 1
|
0.0488090050142807
|
5166
|
105
|
40
|
0.380952380952381
|
0.00774293457220286
|
|
TF
|
TF:M11636
|
Factor: Sox-12; motif: ACCGAACAATR
|
0.0488090050142807
|
545
|
13
|
3
|
0.230769230769231
|
0.0055045871559633
|
|
TF
|
TF:M10529_1
|
Factor: Sp1; motif: RGGGMGGRGSNGGGG; match class: 1
|
0.0488090050142807
|
2816
|
114
|
27
|
0.236842105263158
|
0.00958806818181818
|
|
TF
|
TF:M07038_1
|
Factor: DBP; motif: TTRCATAANN; match class: 1
|
0.0488102479526796
|
772
|
54
|
7
|
0.12962962962963
|
0.00906735751295337
|
|
TF
|
TF:M10816
|
Factor: HOXC11; motif: NGYMATWAANN
|
0.0488102479526796
|
5209
|
10
|
7
|
0.7
|
0.00134382799001728
|
|
TF
|
TF:M11404_1
|
Factor: Fli-1; motif: NACCGGAARTN; match class: 1
|
0.0491257044330771
|
3171
|
69
|
20
|
0.289855072463768
|
0.00630715862503942
|
|
TF
|
TF:M10600
|
Factor: Dlx-7; motif: NTCRTTAN
|
0.049240233456428
|
7477
|
13
|
10
|
0.769230769230769
|
0.0013374348000535
|
|
TF
|
TF:M03988_1
|
Factor: FLI1; motif: ACCGGAARTN; match class: 1
|
0.0496793206215532
|
2950
|
69
|
19
|
0.27536231884058
|
0.00644067796610169
|
|
TF
|
TF:M10112
|
Factor: Miz-1; motif: NNRGGWGGGGGAGGGGMRR
|
0.0496793206215532
|
8711
|
107
|
61
|
0.570093457943925
|
0.00700264033980025
|
|
TF
|
TF:M00195
|
Factor: Oct-1; motif: NNNNATGCAAATNAN
|
0.0497906659861328
|
3659
|
5
|
4
|
0.8
|
0.00109319486198415
|
|
TF
|
TF:M09966
|
Factor: Kaiso; motif: SARNYCTCGCGAGAN
|
0.0498958420797467
|
2356
|
142
|
28
|
0.197183098591549
|
0.0118845500848896
|
|
WP
|
WP:WP111
|
Electron Transport Chain (OXPHOS system in mitochondria)
|
7.33288388591815e-15
|
105
|
13
|
10
|
0.769230769230769
|
0.0952380952380952
|
|
WP
|
WP:WP623
|
Oxidative phosphorylation
|
9.61823717070128e-10
|
62
|
25
|
8
|
0.32
|
0.129032258064516
|
|
WP
|
WP:WP4324
|
Mitochondrial complex I assembly model OXPHOS system
|
1.03331525375521e-06
|
56
|
25
|
6
|
0.24
|
0.107142857142857
|
|
WP
|
WP:WP4922
|
Mitochondrial CIV Assembly
|
1.420145226772e-05
|
35
|
4
|
3
|
0.75
|
0.0857142857142857
|
|
WP
|
WP:WP4396
|
Nonalcoholic fatty liver disease
|
0.000939192295579555
|
159
|
4
|
3
|
0.75
|
0.0188679245283019
|