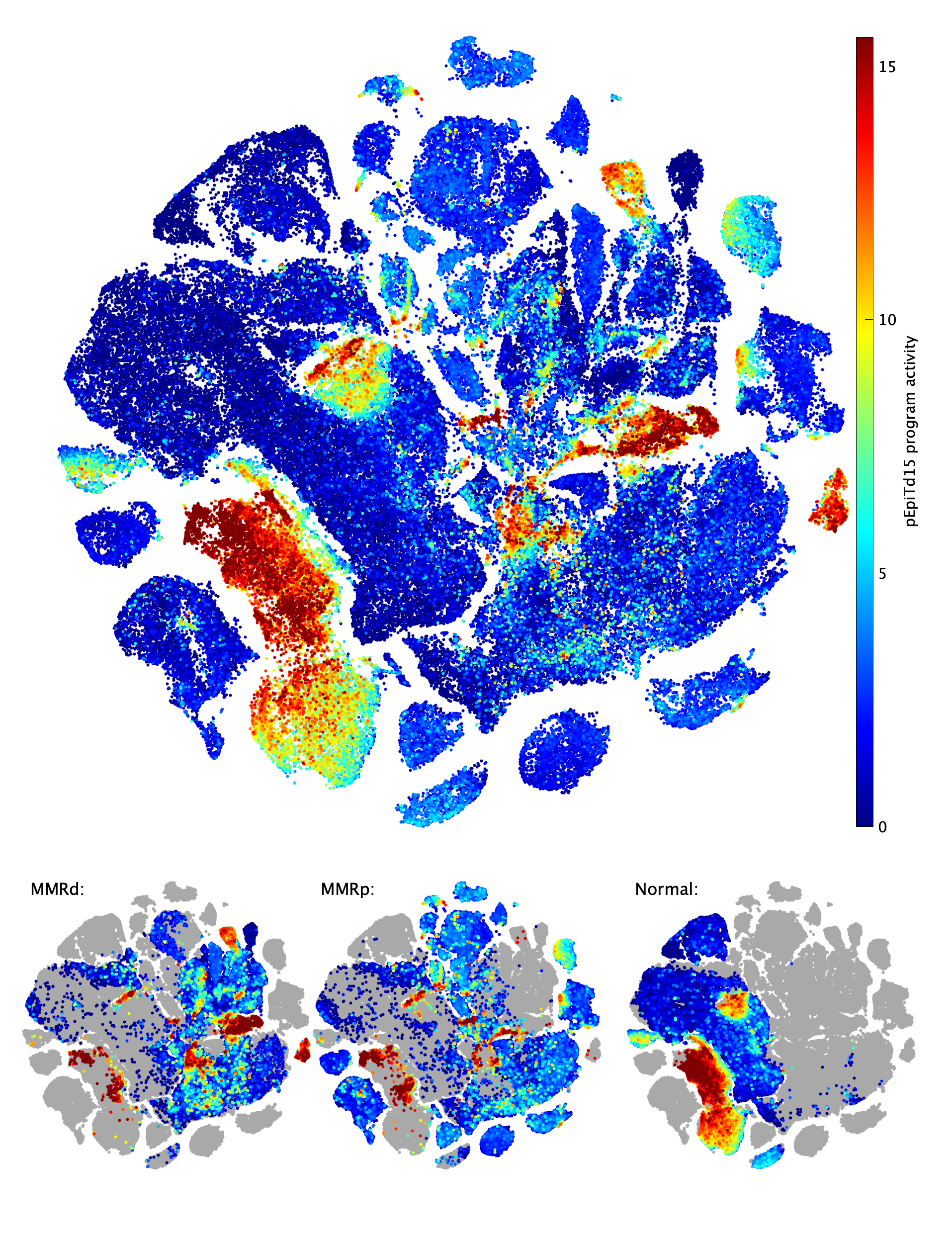
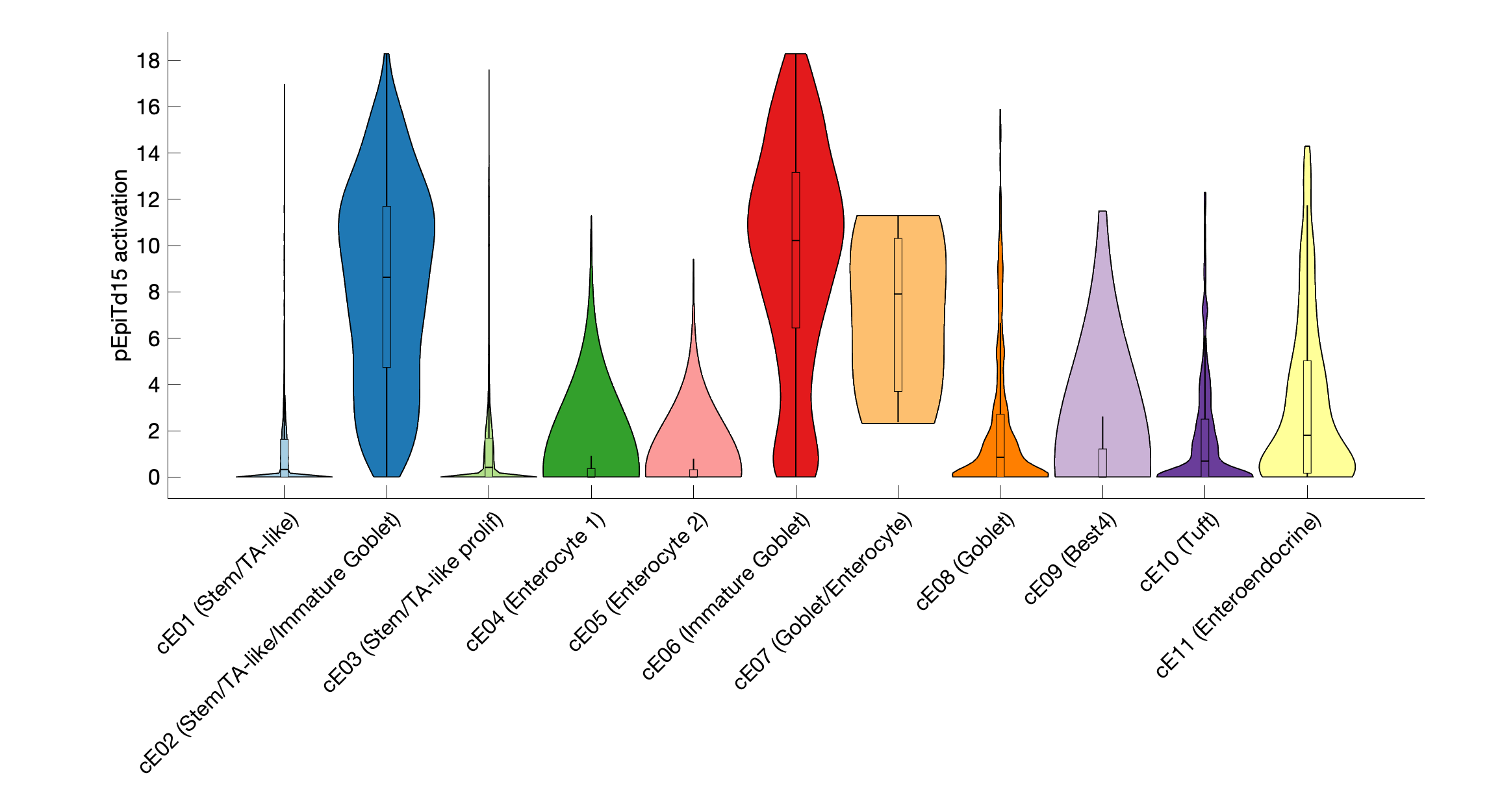
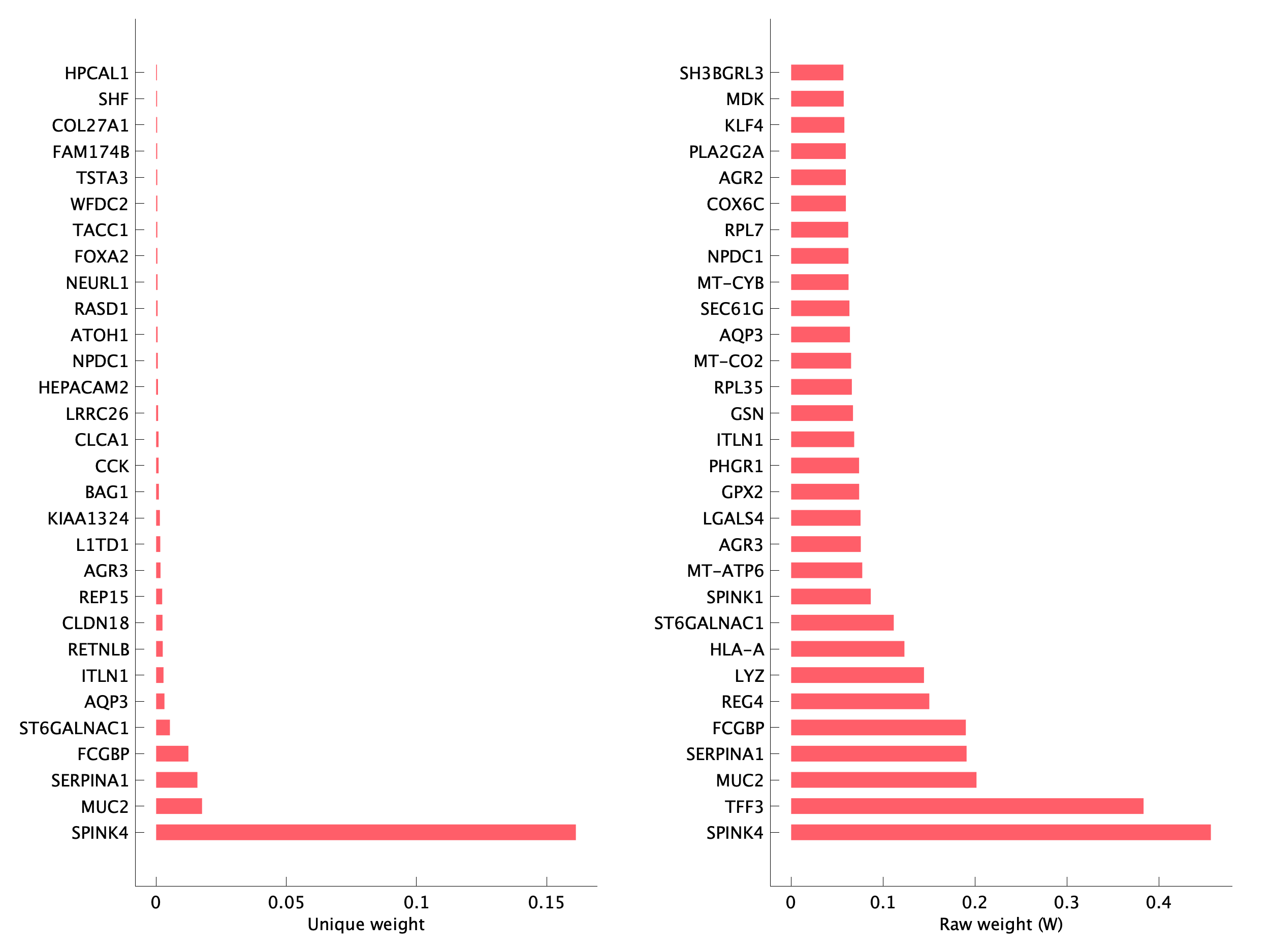
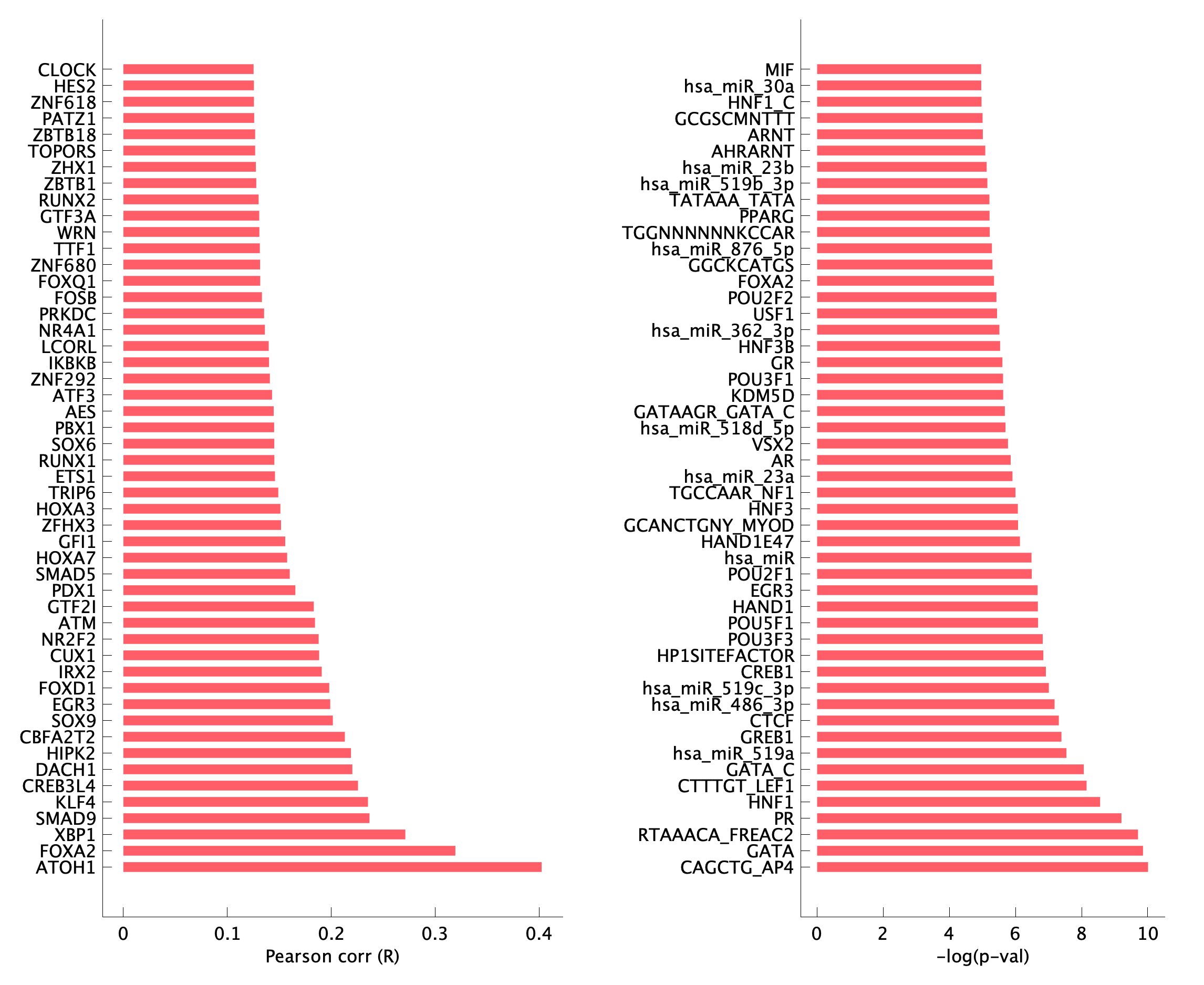
|
GO:CC
|
GO:0043195
|
terminal bouton
|
0.00308112995446502
|
57
|
57
|
4
|
0.0701754385964912
|
0.0701754385964912
|
|
GO:CC
|
GO:0005615
|
extracellular space
|
0.00308112995446502
|
3594
|
76
|
30
|
0.394736842105263
|
0.00834724540901502
|
|
GO:CC
|
GO:0005576
|
extracellular region
|
0.00308112995446502
|
4567
|
73
|
34
|
0.465753424657534
|
0.00744471206481279
|
|
GO:CC
|
GO:0099503
|
secretory vesicle
|
0.0153866845077745
|
1020
|
57
|
11
|
0.192982456140351
|
0.0107843137254902
|
|
GO:CC
|
GO:0012505
|
endomembrane system
|
0.0153866845077745
|
4640
|
113
|
45
|
0.398230088495575
|
0.00969827586206897
|
|
GO:CC
|
GO:0030141
|
secretory granule
|
0.0162288003798527
|
851
|
49
|
9
|
0.183673469387755
|
0.0105757931844888
|
|
GO:CC
|
GO:0016528
|
sarcoplasm
|
0.0179225598244493
|
79
|
88
|
4
|
0.0454545454545455
|
0.0506329113924051
|
|
GO:CC
|
GO:0030054
|
cell junction
|
0.0179225598244493
|
2105
|
70
|
18
|
0.257142857142857
|
0.00855106888361045
|
|
GO:CC
|
GO:0071944
|
cell periphery
|
0.0179225598244493
|
6169
|
71
|
37
|
0.52112676056338
|
0.005997730588426
|
|
GO:CC
|
GO:0043679
|
axon terminus
|
0.0184876544418409
|
127
|
57
|
4
|
0.0701754385964912
|
0.031496062992126
|
|
GO:CC
|
GO:0070382
|
exocytic vesicle
|
0.0184876544418409
|
229
|
57
|
5
|
0.087719298245614
|
0.0218340611353712
|
|
GO:CC
|
GO:0031982
|
vesicle
|
0.021684075753874
|
4055
|
70
|
27
|
0.385714285714286
|
0.00665844636251541
|
|
GO:CC
|
GO:0044306
|
neuron projection terminus
|
0.022389956606804
|
144
|
57
|
4
|
0.0701754385964912
|
0.0277777777777778
|
|
GO:CC
|
GO:0070062
|
extracellular exosome
|
0.0250499338758002
|
2176
|
61
|
16
|
0.262295081967213
|
0.00735294117647059
|
|
GO:CC
|
GO:0062023
|
collagen-containing extracellular matrix
|
0.0320034353230836
|
421
|
121
|
9
|
0.0743801652892562
|
0.0213776722090261
|
|
GO:CC
|
GO:0043230
|
extracellular organelle
|
0.0320034353230836
|
2263
|
61
|
16
|
0.262295081967213
|
0.0070702607158639
|
|
GO:CC
|
GO:1903561
|
extracellular vesicle
|
0.0320034353230836
|
2261
|
61
|
16
|
0.262295081967213
|
0.0070765148164529
|
|
GO:CC
|
GO:0098793
|
presynapse
|
0.0327316000993562
|
521
|
64
|
7
|
0.109375
|
0.0134357005758157
|
|
GO:CC
|
GO:0030133
|
transport vesicle
|
0.0364265973685813
|
414
|
105
|
8
|
0.0761904761904762
|
0.0193236714975845
|
|
GO:CC
|
GO:0005886
|
plasma membrane
|
0.0382838347606116
|
5681
|
71
|
33
|
0.464788732394366
|
0.00580883647245203
|
|
GO:CC
|
GO:0097708
|
intracellular vesicle
|
0.0382838347606116
|
2428
|
70
|
18
|
0.257142857142857
|
0.00741350906095552
|
|
GO:CC
|
GO:0031410
|
cytoplasmic vesicle
|
0.0382838347606116
|
2424
|
70
|
18
|
0.257142857142857
|
0.00742574257425743
|
|
GO:CC
|
GO:0030312
|
external encapsulating structure
|
0.0451454204255146
|
563
|
121
|
10
|
0.0826446280991736
|
0.0177619893428064
|
|
GO:CC
|
GO:0031012
|
extracellular matrix
|
0.0451454204255146
|
562
|
121
|
10
|
0.0826446280991736
|
0.0177935943060498
|
|
GO:CC
|
GO:0008021
|
synaptic vesicle
|
0.0477422001124378
|
209
|
57
|
4
|
0.0701754385964912
|
0.0191387559808612
|
|
GO:CC
|
GO:0150034
|
distal axon
|
0.0489536873301801
|
308
|
64
|
5
|
0.078125
|
0.0162337662337662
|
|
GO:CC
|
GO:0030672
|
synaptic vesicle membrane
|
0.0494078411510804
|
111
|
57
|
3
|
0.0526315789473684
|
0.027027027027027
|
|
GO:CC
|
GO:0099501
|
exocytic vesicle membrane
|
0.0494078411510804
|
111
|
57
|
3
|
0.0526315789473684
|
0.027027027027027
|
|
GO:CC
|
GO:0016529
|
sarcoplasmic reticulum
|
0.0494078411510804
|
70
|
88
|
3
|
0.0340909090909091
|
0.0428571428571429
|
|
GO:MF
|
GO:0005229
|
intracellular calcium activated chloride channel activity
|
0.00264490584721354
|
16
|
52
|
3
|
0.0576923076923077
|
0.1875
|
|
GO:MF
|
GO:0061778
|
intracellular chloride channel activity
|
0.00264490584721354
|
16
|
52
|
3
|
0.0576923076923077
|
0.1875
|
|
GO:MF
|
GO:0022839
|
ion gated channel activity
|
0.00748752798770591
|
43
|
87
|
4
|
0.0459770114942529
|
0.0930232558139535
|
|
GO:MF
|
GO:0008509
|
anion transmembrane transporter activity
|
0.0157398310820222
|
464
|
127
|
11
|
0.0866141732283465
|
0.0237068965517241
|
|
GO:MF
|
GO:0015108
|
chloride transmembrane transporter activity
|
0.0157398310820222
|
99
|
57
|
4
|
0.0701754385964912
|
0.0404040404040404
|
|
GO:MF
|
GO:0004713
|
protein tyrosine kinase activity
|
0.0239916541388995
|
144
|
96
|
5
|
0.0520833333333333
|
0.0347222222222222
|
|
GO:MF
|
GO:0004252
|
serine-type endopeptidase activity
|
0.0239916541388995
|
171
|
123
|
6
|
0.0487804878048781
|
0.0350877192982456
|
|
GO:MF
|
GO:0015175
|
neutral amino acid transmembrane transporter activity
|
0.0239916541388995
|
34
|
102
|
3
|
0.0294117647058824
|
0.0882352941176471
|
|
GO:MF
|
GO:0015103
|
inorganic anion transmembrane transporter activity
|
0.0239916541388995
|
142
|
57
|
4
|
0.0701754385964912
|
0.028169014084507
|
|
GO:MF
|
GO:0015026
|
coreceptor activity
|
0.0239916541388995
|
47
|
68
|
3
|
0.0441176470588235
|
0.0638297872340425
|
|
GO:MF
|
GO:0005509
|
calcium ion binding
|
0.0239916541388995
|
721
|
128
|
13
|
0.1015625
|
0.0180305131761442
|
|
GO:MF
|
GO:0005254
|
chloride channel activity
|
0.0239916541388995
|
73
|
52
|
3
|
0.0576923076923077
|
0.0410958904109589
|
|
GO:MF
|
GO:0005253
|
anion channel activity
|
0.025407507891247
|
85
|
52
|
3
|
0.0576923076923077
|
0.0352941176470588
|
|
GO:MF
|
GO:0008236
|
serine-type peptidase activity
|
0.025407507891247
|
189
|
123
|
6
|
0.0487804878048781
|
0.0317460317460317
|
|
GO:MF
|
GO:0015318
|
inorganic molecular entity transmembrane transporter activity
|
0.025407507891247
|
831
|
127
|
14
|
0.110236220472441
|
0.0168471720818291
|
|
GO:MF
|
GO:0017171
|
serine hydrolase activity
|
0.025407507891247
|
193
|
123
|
6
|
0.0487804878048781
|
0.0310880829015544
|
|
GO:MF
|
GO:0022857
|
transmembrane transporter activity
|
0.0265591863285892
|
1075
|
102
|
14
|
0.137254901960784
|
0.0130232558139535
|
|
GO:MF
|
GO:0015267
|
channel activity
|
0.0265591863285892
|
484
|
52
|
6
|
0.115384615384615
|
0.012396694214876
|
|
GO:MF
|
GO:0022803
|
passive transmembrane transporter activity
|
0.0265591863285892
|
485
|
52
|
6
|
0.115384615384615
|
0.0123711340206186
|
|
GO:MF
|
GO:0015075
|
ion transmembrane transporter activity
|
0.0265591863285892
|
956
|
102
|
13
|
0.127450980392157
|
0.0135983263598326
|
|
GO:MF
|
GO:0140096
|
catalytic activity, acting on a protein
|
0.0265591863285892
|
2332
|
71
|
18
|
0.253521126760563
|
0.00771869639794168
|
|
GO:MF
|
GO:0001784
|
phosphotyrosine residue binding
|
0.0277108071912661
|
44
|
114
|
3
|
0.0263157894736842
|
0.0681818181818182
|
|
GO:MF
|
GO:0022836
|
gated channel activity
|
0.0280701371755331
|
341
|
52
|
5
|
0.0961538461538462
|
0.0146627565982405
|
|
GO:MF
|
GO:0042887
|
amide transmembrane transporter activity
|
0.029830147870947
|
58
|
90
|
3
|
0.0333333333333333
|
0.0517241379310345
|
|
GO:MF
|
GO:0015179
|
L-amino acid transmembrane transporter activity
|
0.0341433057056373
|
56
|
102
|
3
|
0.0294117647058824
|
0.0535714285714286
|
|
GO:MF
|
GO:0004175
|
endopeptidase activity
|
0.0349174103406732
|
450
|
63
|
6
|
0.0952380952380952
|
0.0133333333333333
|
|
GO:MF
|
GO:0045309
|
protein phosphorylated amino acid binding
|
0.0381340935320641
|
56
|
114
|
3
|
0.0263157894736842
|
0.0535714285714286
|
|
GO:MF
|
GO:1901618
|
organic hydroxy compound transmembrane transporter activity
|
0.0381340935320641
|
50
|
127
|
3
|
0.0236220472440945
|
0.06
|
|
GO:MF
|
GO:0005215
|
transporter activity
|
0.0385771505750855
|
1185
|
102
|
14
|
0.137254901960784
|
0.0118143459915612
|
|
GO:MF
|
GO:0005201
|
extracellular matrix structural constituent
|
0.038826536346787
|
171
|
121
|
5
|
0.0413223140495868
|
0.0292397660818713
|
|
GO:MF
|
GO:0003713
|
transcription coactivator activity
|
0.0395237164026719
|
280
|
47
|
4
|
0.0851063829787234
|
0.0142857142857143
|
|
GO:MF
|
GO:0008233
|
peptidase activity
|
0.0397152188564532
|
646
|
63
|
7
|
0.111111111111111
|
0.0108359133126935
|
|
GO:MF
|
GO:0005216
|
ion channel activity
|
0.044394332989933
|
435
|
52
|
5
|
0.0961538461538462
|
0.0114942528735632
|
|
GO:MF
|
GO:0098772
|
molecular function regulator
|
0.0452144053462171
|
2363
|
24
|
8
|
0.333333333333333
|
0.00338552687261955
|
|
GO:MF
|
GO:0019838
|
growth factor binding
|
0.0487390304748893
|
138
|
107
|
4
|
0.0373831775700935
|
0.0289855072463768
|
|
GO:MF
|
GO:0019199
|
transmembrane receptor protein kinase activity
|
0.0496830833541279
|
82
|
96
|
3
|
0.03125
|
0.0365853658536585
|
|
GO:MF
|
GO:0015171
|
amino acid transmembrane transporter activity
|
0.0496830833541279
|
78
|
102
|
3
|
0.0294117647058824
|
0.0384615384615385
|
|
REAC
|
REAC:R-HSA-2672351
|
Stimuli-sensing channels
|
0.0271568490940423
|
105
|
71
|
6
|
0.0845070422535211
|
0.0571428571428571
|
|
TF
|
TF:M00176
|
Factor: AP-4; motif: CWCAGCTGGN
|
0.0429909572628257
|
2528
|
70
|
21
|
0.3
|
0.00830696202531645
|
|
TF
|
TF:M10097_1
|
Factor: AP-4; motif: YCAGCTGNKN; match class: 1
|
0.0429909572628257
|
1554
|
62
|
15
|
0.241935483870968
|
0.00965250965250965
|
|
TF
|
TF:M04797
|
Factor: Egr-1; motif: CCGCCCMCG
|
0.0429909572628257
|
5116
|
120
|
50
|
0.416666666666667
|
0.00977326035965598
|
|
TF
|
TF:M12096_1
|
Factor: ZNF704; motif: NRCCGGCCGGYN; match class: 1
|
0.0429909572628257
|
80
|
111
|
5
|
0.045045045045045
|
0.0625
|
|
TF
|
TF:M12096
|
Factor: ZNF704; motif: NRCCGGCCGGYN
|
0.0429909572628257
|
80
|
111
|
5
|
0.045045045045045
|
0.0625
|
|
TF
|
TF:M09727_1
|
Factor: GEMIN3; motif: NCWGGRARRGRGNGNG; match class: 1
|
0.0429909572628257
|
3210
|
128
|
38
|
0.296875
|
0.0118380062305296
|
|
TF
|
TF:M06089
|
Factor: ZNF26; motif: NAMTCAAGSGTA
|
0.0429909572628257
|
30
|
61
|
3
|
0.0491803278688525
|
0.1
|
|
TF
|
TF:M11728
|
Factor: NF-1C; motif: NTTGGCNNNNTGCCARN
|
0.0429909572628257
|
2634
|
16
|
9
|
0.5625
|
0.00341685649202733
|
|
TF
|
TF:M04152
|
Factor: TFAP2C; motif: NGCCCNNRGGCA
|
0.0429909572628257
|
7738
|
128
|
74
|
0.578125
|
0.00956319462393383
|
|
TF
|
TF:M09973_1
|
Factor: CPBP; motif: GNNRGGGHGGGGNNGGGRN; match class: 1
|
0.0429909572628257
|
6607
|
129
|
64
|
0.496124031007752
|
0.00968669592856062
|
|
TF
|
TF:M04811_1
|
Factor: RelA-p65; motif: BCWGGGRANNK; match class: 1
|
0.0429909572628257
|
1834
|
10
|
6
|
0.6
|
0.00327153762268266
|
|
TF
|
TF:M01196_1
|
Factor: CTF1; motif: TGGCASCNNGCCAA; match class: 1
|
0.0429909572628257
|
1572
|
16
|
7
|
0.4375
|
0.0044529262086514
|
|
TF
|
TF:M07044
|
Factor: HNF-1alpha; motif: NGNNAAWNATTAACNN
|
0.0429909572628257
|
1678
|
11
|
6
|
0.545454545454545
|
0.00357568533969011
|
|
TF
|
TF:M07611_1
|
Factor: p54NRB; motif: GRNNNMGGATGRMNNCGGA; match class: 1
|
0.0429909572628257
|
1645
|
38
|
12
|
0.315789473684211
|
0.00729483282674772
|
|
TF
|
TF:M11124
|
Factor: Olig3; motif: ANCAKMTGTT
|
0.0429909572628257
|
1103
|
20
|
7
|
0.35
|
0.00634632819582956
|
|
TF
|
TF:M10002
|
Factor: NF-1C; motif: NYTGGCNNYNNGCCARN
|
0.0429909572628257
|
6760
|
71
|
41
|
0.577464788732394
|
0.00606508875739645
|
|
TF
|
TF:M07338
|
Factor: Sox-10; motif: ACAAWGNN
|
0.0429909572628257
|
1695
|
7
|
5
|
0.714285714285714
|
0.00294985250737463
|
|
TF
|
TF:M07354_1
|
Factor: Egr-1; motif: GCGGGGGCGG; match class: 1
|
0.0464502696539512
|
3561
|
126
|
40
|
0.317460317460317
|
0.011232799775344
|
|
TF
|
TF:M04146_1
|
Factor: TFAP2A; motif: YGCCCNNRGGCN; match class: 1
|
0.0492557056831396
|
3800
|
116
|
39
|
0.336206896551724
|
0.0102631578947368
|
|
TF
|
TF:M11728_1
|
Factor: NF-1C; motif: NTTGGCNNNNTGCCARN; match class: 1
|
0.0492557056831396
|
1129
|
16
|
6
|
0.375
|
0.00531443755535872
|
|
TF
|
TF:M09640
|
Factor: NF-1C; motif: NYYTGGCWNNNNKCCMN
|
0.0492557056831396
|
2807
|
8
|
6
|
0.75
|
0.0021375133594585
|
|
TF
|
TF:M03906
|
Factor: HIC2; motif: RTGCCCRNN
|
0.0492557056831396
|
3458
|
7
|
6
|
0.857142857142857
|
0.00173510699826489
|
|
TF
|
TF:M11208_1
|
Factor: AP-4; motif: AWCAGCTGWT; match class: 1
|
0.0499010485213191
|
3864
|
28
|
14
|
0.5
|
0.0036231884057971
|
|
TF
|
TF:M04146
|
Factor: TFAP2A; motif: YGCCCNNRGGCN
|
0.0499010485213191
|
6552
|
116
|
57
|
0.491379310344828
|
0.0086996336996337
|
|
TF
|
TF:M04106_1
|
Factor: RUNX2; motif: NRACCGCAAACCGCAN; match class: 1
|
0.0499010485213191
|
8372
|
127
|
74
|
0.582677165354331
|
0.00883898709985667
|
|
TF
|
TF:M01860
|
Factor: AP-4; motif: NCAGCTGYNGNCN
|
0.0499010485213191
|
5995
|
110
|
51
|
0.463636363636364
|
0.0085070892410342
|
|
TF
|
TF:M08961
|
Factor: PPARGAMMA; motif: NWNTRGGTYANN
|
0.0499010485213191
|
5433
|
11
|
9
|
0.818181818181818
|
0.00165654334621756
|
|
TF
|
TF:M01196
|
Factor: CTF1; motif: TGGCASCNNGCCAA
|
0.0499010485213191
|
4864
|
16
|
11
|
0.6875
|
0.00226151315789474
|
|
TF
|
TF:M00034_1
|
Factor: p53; motif: GGACATGCCCGGGCATGTCY; match class: 1
|
0.0499010485213191
|
2646
|
6
|
5
|
0.833333333333333
|
0.0018896447467876
|
|
TF
|
TF:M11729
|
Factor: NF-1C; motif: NTTGGCNNNNTGCCARN
|
0.0499010485213191
|
1738
|
16
|
7
|
0.4375
|
0.00402761795166858
|
|
TF
|
TF:M10432_1
|
Factor: MAZ; motif: GGGMGGGGS; match class: 1
|
0.0499010485213191
|
4365
|
128
|
46
|
0.359375
|
0.0105383734249714
|
|
TF
|
TF:M11097
|
Factor: Max; motif: MCATGTGNNNNMCATGTGN
|
0.0499010485213191
|
618
|
5
|
3
|
0.6
|
0.00485436893203883
|
|
TF
|
TF:M12160_1
|
Factor: KLF15; motif: RCCMCRCCCMCN; match class: 1
|
0.0499010485213191
|
8031
|
129
|
73
|
0.565891472868217
|
0.00908977711368447
|
|
TF
|
TF:M01837
|
Factor: FKLF; motif: BGGGNGGVMD
|
0.0499010485213191
|
5442
|
128
|
54
|
0.421875
|
0.00992282249173098
|
|
TF
|
TF:M00915_1
|
Factor: AP-2; motif: SNNNCCNCAGGCN; match class: 1
|
0.0499010485213191
|
4551
|
126
|
47
|
0.373015873015873
|
0.010327400571303
|
|
TF
|
TF:M00257
|
Factor: RREB-1; motif: CCCCAAACMMCCCC
|
0.0499010485213191
|
10516
|
129
|
89
|
0.689922480620155
|
0.00846329402814758
|
|
TF
|
TF:M01651
|
Factor: P53; motif: RGRCATGYCYRGRCATGYYY
|
0.0499010485213191
|
2163
|
7
|
5
|
0.714285714285714
|
0.00231160425335183
|