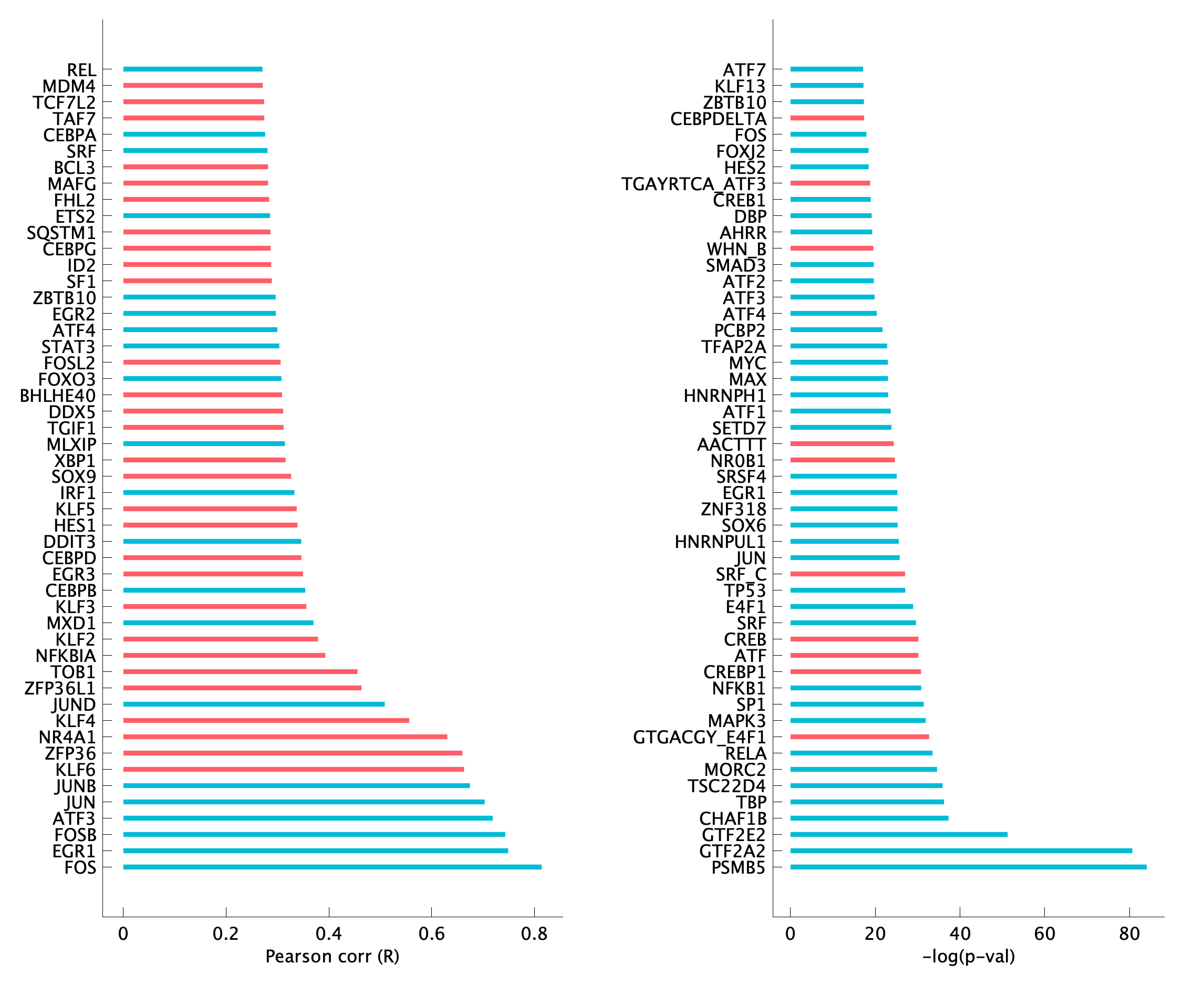
|
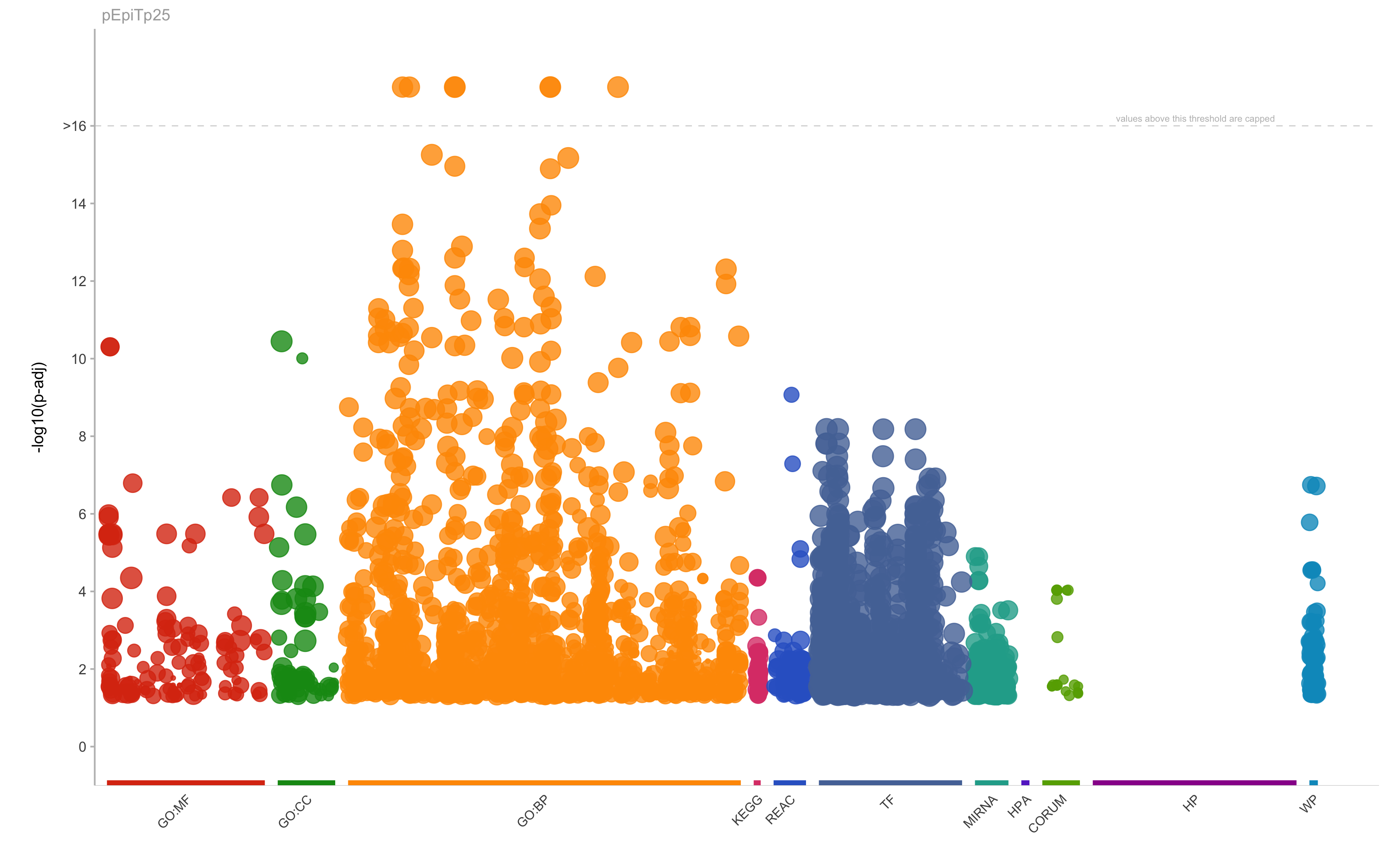
GO:BP
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
8.10294361970785e-19
|
3083
|
100
|
60
|
0.6
|
0.0194615634122608
|
|
GO:BP
|
GO:0031325
|
positive regulation of cellular metabolic process
|
8.10294361970785e-19
|
3274
|
101
|
62
|
0.613861386138614
|
0.0189370800244349
|
|
GO:BP
|
GO:0031323
|
regulation of cellular metabolic process
|
6.71280677220206e-18
|
6269
|
101
|
81
|
0.801980198019802
|
0.0129207210081353
|
|
GO:BP
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
1.62709376138569e-17
|
5848
|
96
|
75
|
0.78125
|
0.0128248974008208
|
|
GO:BP
|
GO:0009893
|
positive regulation of metabolic process
|
1.62709376138569e-17
|
3783
|
101
|
64
|
0.633663366336634
|
0.0169177901136664
|
|
GO:BP
|
GO:0080090
|
regulation of primary metabolic process
|
1.62709376138569e-17
|
6053
|
101
|
79
|
0.782178217821782
|
0.013051379481249
|
|
GO:BP
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
1.62709376138569e-17
|
3483
|
92
|
58
|
0.630434782608696
|
0.0166523112259546
|
|
GO:BP
|
GO:0019222
|
regulation of metabolic process
|
5.57347444489069e-16
|
7148
|
101
|
83
|
0.821782178217822
|
0.011611639619474
|
|
GO:BP
|
GO:0060255
|
regulation of macromolecule metabolic process
|
6.73247668389968e-16
|
6603
|
96
|
77
|
0.802083333333333
|
0.0116613660457368
|
|
GO:BP
|
GO:0031324
|
negative regulation of cellular metabolic process
|
1.10010102561983e-15
|
2719
|
96
|
51
|
0.53125
|
0.0187568959176168
|
|
GO:BP
|
GO:0051172
|
negative regulation of nitrogen compound metabolic process
|
1.27368675631716e-15
|
2518
|
89
|
47
|
0.528089887640449
|
0.0186656076250993
|
|
GO:BP
|
GO:0051254
|
positive regulation of RNA metabolic process
|
1.11591500459587e-14
|
1714
|
113
|
44
|
0.389380530973451
|
0.0256709451575263
|
|
GO:BP
|
GO:0048522
|
positive regulation of cellular process
|
1.8768865856868e-14
|
5757
|
101
|
73
|
0.722772277227723
|
0.01268021538996
|
|
GO:BP
|
GO:0009889
|
regulation of biosynthetic process
|
3.45059293910234e-14
|
4361
|
100
|
63
|
0.63
|
0.014446227929374
|
|
GO:BP
|
GO:0048518
|
positive regulation of biological process
|
4.44140032516426e-14
|
6363
|
101
|
76
|
0.752475247524752
|
0.0119440515480119
|
|
GO:BP
|
GO:0032502
|
developmental process
|
1.28283092986639e-13
|
6628
|
96
|
74
|
0.770833333333333
|
0.0111647555823778
|
|
GO:BP
|
GO:0009892
|
negative regulation of metabolic process
|
1.61765582286696e-13
|
3479
|
96
|
54
|
0.5625
|
0.0155217016384018
|
|
GO:BP
|
GO:0031326
|
regulation of cellular biosynthetic process
|
2.53459707041719e-13
|
4273
|
100
|
61
|
0.61
|
0.0142756845307746
|
|
GO:BP
|
GO:0045935
|
positive regulation of nucleobase-containing compound metabolic process
|
2.53459707041719e-13
|
1887
|
113
|
44
|
0.389380530973451
|
0.023317435082141
|
|
GO:BP
|
GO:0045944
|
positive regulation of transcription by RNA polymerase II
|
4.37509472567783e-13
|
1197
|
124
|
37
|
0.298387096774194
|
0.0309106098579783
|
|
GO:BP
|
GO:0010033
|
response to organic substance
|
4.61876116557389e-13
|
3412
|
45
|
33
|
0.733333333333333
|
0.00967174677608441
|
|
GO:BP
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
4.6952756457341e-13
|
3241
|
89
|
49
|
0.550561797752809
|
0.0151187904967603
|
|
GO:BP
|
GO:0009890
|
negative regulation of biosynthetic process
|
4.6952756457341e-13
|
1740
|
95
|
38
|
0.4
|
0.0218390804597701
|
|
GO:BP
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
4.92715035760751e-13
|
4071
|
100
|
59
|
0.59
|
0.0144927536231884
|
|
GO:BP
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
6.79625501973367e-13
|
4102
|
100
|
59
|
0.59
|
0.0143832276938079
|
|
GO:BP
|
GO:0070887
|
cellular response to chemical stimulus
|
7.58369819968612e-13
|
3393
|
68
|
42
|
0.617647058823529
|
0.0123784261715296
|
|
GO:BP
|
GO:0048523
|
negative regulation of cellular process
|
8.91283713044417e-13
|
5139
|
96
|
64
|
0.666666666666667
|
0.0124537847830317
|
|
GO:BP
|
GO:2000113
|
negative regulation of cellular macromolecule biosynthetic process
|
1.19121120218844e-12
|
1628
|
89
|
35
|
0.393258426966292
|
0.0214987714987715
|
|
GO:BP
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
1.28956451842185e-12
|
1707
|
95
|
37
|
0.389473684210526
|
0.0216754540128881
|
|
GO:BP
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
1.36251250690744e-12
|
1639
|
89
|
35
|
0.393258426966292
|
0.0213544844417328
|
|
GO:BP
|
GO:0048856
|
anatomical structure development
|
2.50248169987028e-12
|
6106
|
96
|
69
|
0.71875
|
0.0113003603013429
|
|
GO:BP
|
GO:0032268
|
regulation of cellular protein metabolic process
|
2.91613499948958e-12
|
2541
|
91
|
43
|
0.472527472527473
|
0.016922471467926
|
|
GO:BP
|
GO:0042221
|
response to chemical
|
2.96460744864144e-12
|
4781
|
46
|
37
|
0.804347826086957
|
0.00773896674335913
|
|
GO:BP
|
GO:0051246
|
regulation of protein metabolic process
|
4.65966772277248e-12
|
2704
|
91
|
44
|
0.483516483516484
|
0.0162721893491124
|
|
GO:BP
|
GO:0010941
|
regulation of cell death
|
4.97161989116163e-12
|
1720
|
89
|
35
|
0.393258426966292
|
0.0203488372093023
|
|
GO:BP
|
GO:0006357
|
regulation of transcription by RNA polymerase II
|
5.11997740750866e-12
|
2555
|
92
|
43
|
0.467391304347826
|
0.0168297455968689
|
|
GO:BP
|
GO:0006366
|
transcription by RNA polymerase II
|
9.03204169160937e-12
|
2723
|
92
|
44
|
0.478260869565217
|
0.0161586485493941
|
|
GO:BP
|
GO:0042981
|
regulation of apoptotic process
|
9.03204169160937e-12
|
1549
|
89
|
33
|
0.370786516853933
|
0.0213040671400904
|
|
GO:BP
|
GO:0051252
|
regulation of RNA metabolic process
|
9.37529042872314e-12
|
3822
|
100
|
55
|
0.55
|
0.0143903715332287
|
|
GO:BP
|
GO:0006915
|
apoptotic process
|
9.84792030050645e-12
|
1990
|
89
|
37
|
0.415730337078652
|
0.0185929648241206
|
|
GO:BP
|
GO:0033554
|
cellular response to stress
|
1.0500388820855e-11
|
2111
|
89
|
38
|
0.426966292134831
|
0.0180009474182852
|
|
GO:BP
|
GO:0048583
|
regulation of response to stimulus
|
1.26389851038806e-11
|
4278
|
85
|
52
|
0.611764705882353
|
0.0121552127162225
|
|
GO:BP
|
GO:0043067
|
regulation of programmed cell death
|
1.44740666135888e-11
|
1582
|
89
|
33
|
0.370786516853933
|
0.0208596713021492
|
|
GO:BP
|
GO:1903508
|
positive regulation of nucleic acid-templated transcription
|
1.54264162651714e-11
|
1617
|
113
|
38
|
0.336283185840708
|
0.0235003092145949
|
|
GO:BP
|
GO:1902680
|
positive regulation of RNA biosynthetic process
|
1.54264162651714e-11
|
1618
|
113
|
38
|
0.336283185840708
|
0.0234857849196539
|
|
GO:BP
|
GO:0045893
|
positive regulation of transcription, DNA-templated
|
1.54264162651714e-11
|
1617
|
113
|
38
|
0.336283185840708
|
0.0235003092145949
|
|
GO:BP
|
GO:0010468
|
regulation of gene expression
|
1.63033787241675e-11
|
5196
|
100
|
64
|
0.64
|
0.0123171670515781
|
|
GO:BP
|
GO:0006950
|
response to stress
|
1.71316526984529e-11
|
4170
|
100
|
57
|
0.57
|
0.0136690647482014
|
|
GO:BP
|
GO:0008219
|
cell death
|
2.01308410763123e-11
|
2292
|
85
|
38
|
0.447058823529412
|
0.0165794066317627
|
|
GO:BP
|
GO:0009891
|
positive regulation of biosynthetic process
|
2.20744138611732e-11
|
2004
|
113
|
42
|
0.371681415929204
|
0.0209580838323353
|
|
GO:BP
|
GO:1903506
|
regulation of nucleic acid-templated transcription
|
2.5292727549087e-11
|
3535
|
100
|
52
|
0.52
|
0.0147100424328147
|
|
GO:BP
|
GO:0006355
|
regulation of transcription, DNA-templated
|
2.5292727549087e-11
|
3534
|
100
|
52
|
0.52
|
0.0147142048670062
|
|
GO:BP
|
GO:2001141
|
regulation of RNA biosynthetic process
|
2.62980423024845e-11
|
3540
|
100
|
52
|
0.52
|
0.0146892655367232
|
|
GO:BP
|
GO:0009628
|
response to abiotic stimulus
|
2.75651007515561e-11
|
1261
|
99
|
31
|
0.313131313131313
|
0.0245836637589215
|
|
GO:BP
|
GO:0019219
|
regulation of nucleobase-containing compound metabolic process
|
2.86855987873363e-11
|
4091
|
100
|
56
|
0.56
|
0.0136885846981178
|
|
GO:BP
|
GO:1901700
|
response to oxygen-containing compound
|
3.60410429087599e-11
|
1739
|
76
|
31
|
0.407894736842105
|
0.0178263369752731
|
|
GO:BP
|
GO:0097659
|
nucleic acid-templated transcription
|
3.84485375622146e-11
|
3715
|
100
|
53
|
0.53
|
0.0142664872139973
|
|
GO:BP
|
GO:0006351
|
transcription, DNA-templated
|
3.84485375622146e-11
|
3714
|
100
|
53
|
0.53
|
0.0142703284868067
|
|
GO:BP
|
GO:0007275
|
multicellular organism development
|
3.84485375622146e-11
|
5613
|
96
|
64
|
0.666666666666667
|
0.0114021022626047
|
|
GO:BP
|
GO:0032774
|
RNA biosynthetic process
|
4.51997021699493e-11
|
3731
|
100
|
53
|
0.53
|
0.0142053068882337
|
|
GO:BP
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
4.72341999189498e-11
|
1967
|
113
|
41
|
0.36283185840708
|
0.0208439247585155
|
|
GO:BP
|
GO:0012501
|
programmed cell death
|
6.24396877339993e-11
|
2143
|
89
|
37
|
0.415730337078652
|
0.0172655156322912
|
|
GO:BP
|
GO:0051248
|
negative regulation of protein metabolic process
|
6.24396877339993e-11
|
1151
|
85
|
27
|
0.317647058823529
|
0.0234578627280626
|
|
GO:BP
|
GO:0044260
|
cellular macromolecule metabolic process
|
9.59652767144884e-11
|
8390
|
96
|
78
|
0.8125
|
0.00929678188319428
|
|
GO:BP
|
GO:0048519
|
negative regulation of biological process
|
1.20968989720151e-10
|
5757
|
96
|
64
|
0.666666666666667
|
0.0111169011638006
|
|
GO:BP
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
1.43307524871494e-10
|
1851
|
113
|
39
|
0.345132743362832
|
0.0210696920583468
|
|
GO:BP
|
GO:0080134
|
regulation of response to stress
|
1.72795417012629e-10
|
1443
|
78
|
28
|
0.358974358974359
|
0.0194040194040194
|
|
GO:BP
|
GO:0071310
|
cellular response to organic substance
|
4.13871215449167e-10
|
2776
|
76
|
37
|
0.486842105263158
|
0.013328530259366
|
|
GO:BP
|
GO:0009719
|
response to endogenous stimulus
|
5.48427185383343e-10
|
1683
|
34
|
19
|
0.558823529411765
|
0.0112893642305407
|
|
GO:BP
|
GO:0032269
|
negative regulation of cellular protein metabolic process
|
6.74523666257509e-10
|
1082
|
85
|
25
|
0.294117647058824
|
0.0231053604436229
|
|
GO:BP
|
GO:0048585
|
negative regulation of response to stimulus
|
6.74523666257509e-10
|
1733
|
120
|
38
|
0.316666666666667
|
0.0219272937103289
|
|
GO:BP
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
6.82269952231071e-10
|
5051
|
95
|
58
|
0.610526315789474
|
0.0114828746782815
|
|
GO:BP
|
GO:0045892
|
negative regulation of transcription, DNA-templated
|
7.40654840256559e-10
|
1337
|
94
|
29
|
0.308510638297872
|
0.0216903515332835
|
|
GO:BP
|
GO:1903507
|
negative regulation of nucleic acid-templated transcription
|
7.57712534329642e-10
|
1339
|
94
|
29
|
0.308510638297872
|
0.0216579536967886
|
|
GO:BP
|
GO:1902679
|
negative regulation of RNA biosynthetic process
|
7.75252843025248e-10
|
1341
|
94
|
29
|
0.308510638297872
|
0.0216256524981357
|
|
GO:BP
|
GO:0030097
|
hemopoiesis
|
8.35567003824232e-10
|
928
|
123
|
28
|
0.227642276422764
|
0.0301724137931034
|
|
GO:BP
|
GO:0051253
|
negative regulation of RNA metabolic process
|
8.35567003824232e-10
|
1445
|
94
|
30
|
0.319148936170213
|
0.0207612456747405
|
|
GO:BP
|
GO:0045934
|
negative regulation of nucleobase-containing compound metabolic process
|
8.52656975675172e-10
|
1548
|
94
|
31
|
0.329787234042553
|
0.0200258397932817
|
|
GO:BP
|
GO:0009059
|
macromolecule biosynthetic process
|
1.06354840827842e-09
|
5114
|
95
|
58
|
0.610526315789474
|
0.0113414157215487
|
|
GO:BP
|
GO:0034654
|
nucleobase-containing compound biosynthetic process
|
1.06354840827842e-09
|
4199
|
100
|
54
|
0.54
|
0.0128602048106692
|
|
GO:BP
|
GO:0035556
|
intracellular signal transduction
|
1.10065193313497e-09
|
2772
|
107
|
45
|
0.420560747663551
|
0.0162337662337662
|
|
GO:BP
|
GO:0000122
|
negative regulation of transcription by RNA polymerase II
|
1.77970633866536e-09
|
901
|
89
|
23
|
0.258426966292135
|
0.025527192008879
|
|
GO:BP
|
GO:0010648
|
negative regulation of cell communication
|
1.88700721606496e-09
|
1468
|
90
|
29
|
0.322222222222222
|
0.0197547683923706
|
|
GO:BP
|
GO:0023057
|
negative regulation of signaling
|
1.91836266886738e-09
|
1471
|
90
|
29
|
0.322222222222222
|
0.0197144799456152
|
|
GO:BP
|
GO:0048534
|
hematopoietic or lymphoid organ development
|
1.91836266886738e-09
|
966
|
123
|
28
|
0.227642276422764
|
0.0289855072463768
|
|
GO:BP
|
GO:0018130
|
heterocycle biosynthetic process
|
1.91836266886738e-09
|
4269
|
100
|
54
|
0.54
|
0.0126493323963457
|
|
GO:BP
|
GO:0019438
|
aromatic compound biosynthetic process
|
2.06538544438845e-09
|
4278
|
100
|
54
|
0.54
|
0.0126227208976157
|
|
GO:BP
|
GO:0045595
|
regulation of cell differentiation
|
2.16263492389312e-09
|
1679
|
123
|
37
|
0.300813008130081
|
0.0220369267421084
|
|
GO:BP
|
GO:0033993
|
response to lipid
|
3.19588517476269e-09
|
918
|
42
|
16
|
0.380952380952381
|
0.0174291938997821
|
|
GO:BP
|
GO:0010646
|
regulation of cell communication
|
3.38005324638083e-09
|
3543
|
90
|
45
|
0.5
|
0.012701100762066
|
|
GO:BP
|
GO:0051716
|
cellular response to stimulus
|
3.72274316037932e-09
|
7781
|
92
|
70
|
0.760869565217391
|
0.00899627297262563
|
|
GO:BP
|
GO:0050794
|
regulation of cellular process
|
4.40433502265377e-09
|
11473
|
92
|
85
|
0.923913043478261
|
0.00740869868386647
|
|
GO:BP
|
GO:0023051
|
regulation of signaling
|
4.59775195746941e-09
|
3578
|
90
|
45
|
0.5
|
0.0125768585802124
|
|
GO:BP
|
GO:0032501
|
multicellular organismal process
|
4.88238375426684e-09
|
7946
|
96
|
73
|
0.760416666666667
|
0.00918701233324943
|
|
GO:BP
|
GO:0009966
|
regulation of signal transduction
|
5.45802413334229e-09
|
3165
|
90
|
42
|
0.466666666666667
|
0.0132701421800948
|
|
GO:BP
|
GO:0002520
|
immune system development
|
5.95667746385647e-09
|
1020
|
123
|
28
|
0.227642276422764
|
0.0274509803921569
|
|
GO:BP
|
GO:0044271
|
cellular nitrogen compound biosynthetic process
|
5.96557136004869e-09
|
5017
|
95
|
56
|
0.589473684210526
|
0.0111620490332868
|
|
GO:BP
|
GO:0016070
|
RNA metabolic process
|
6.44831660384552e-09
|
4710
|
95
|
54
|
0.568421052631579
|
0.0114649681528662
|
|
GO:BP
|
GO:1901362
|
organic cyclic compound biosynthetic process
|
8.01242271212997e-09
|
4439
|
100
|
54
|
0.54
|
0.0121649020049561
|
|
GO:BP
|
GO:0010467
|
gene expression
|
9.36223933612702e-09
|
6179
|
100
|
65
|
0.65
|
0.0105195015374656
|
|
GO:BP
|
GO:0050793
|
regulation of developmental process
|
9.80318377268368e-09
|
2605
|
96
|
39
|
0.40625
|
0.0149712092130518
|
|
GO:BP
|
GO:0035914
|
skeletal muscle cell differentiation
|
1.01786221700483e-08
|
68
|
11
|
5
|
0.454545454545455
|
0.0735294117647059
|
|
GO:BP
|
GO:0062197
|
cellular response to chemical stress
|
1.01807102886845e-08
|
365
|
120
|
17
|
0.141666666666667
|
0.0465753424657534
|
|
GO:BP
|
GO:0048513
|
animal organ development
|
1.01807102886845e-08
|
3672
|
96
|
47
|
0.489583333333333
|
0.0127995642701525
|
|
GO:BP
|
GO:0043170
|
macromolecule metabolic process
|
1.02589864801034e-08
|
9796
|
96
|
81
|
0.84375
|
0.00826868109432421
|
|
GO:BP
|
GO:0006468
|
protein phosphorylation
|
1.16550846586123e-08
|
1668
|
114
|
34
|
0.298245614035088
|
0.0203836930455636
|
|
GO:BP
|
GO:0006979
|
response to oxidative stress
|
1.19021971275834e-08
|
466
|
51
|
13
|
0.254901960784314
|
0.0278969957081545
|
|
GO:BP
|
GO:0048731
|
system development
|
1.25358586390345e-08
|
5052
|
96
|
56
|
0.583333333333333
|
0.0110847189231987
|
|
GO:BP
|
GO:0014070
|
response to organic cyclic compound
|
1.28091384132133e-08
|
967
|
23
|
12
|
0.521739130434783
|
0.0124095139607032
|
|
GO:BP
|
GO:0043066
|
negative regulation of apoptotic process
|
1.35844875356084e-08
|
921
|
89
|
22
|
0.247191011235955
|
0.0238870792616721
|
|
GO:BP
|
GO:0070848
|
response to growth factor
|
1.45939115807698e-08
|
759
|
34
|
13
|
0.382352941176471
|
0.0171277997364954
|
|
GO:BP
|
GO:0051726
|
regulation of cell cycle
|
1.64476045055031e-08
|
1236
|
88
|
25
|
0.284090909090909
|
0.0202265372168285
|
|
GO:BP
|
GO:0007154
|
cell communication
|
1.6572221250882e-08
|
6829
|
92
|
64
|
0.695652173913043
|
0.009371796749158
|
|
GO:BP
|
GO:1901701
|
cellular response to oxygen-containing compound
|
1.72639182957113e-08
|
1227
|
76
|
23
|
0.302631578947368
|
0.0187449062754686
|
|
GO:BP
|
GO:1903706
|
regulation of hemopoiesis
|
1.77217243840831e-08
|
424
|
123
|
18
|
0.146341463414634
|
0.0424528301886792
|
|
GO:BP
|
GO:0030154
|
cell differentiation
|
1.82623472444414e-08
|
4350
|
101
|
53
|
0.524752475247525
|
0.012183908045977
|
|
GO:BP
|
GO:0043069
|
negative regulation of programmed cell death
|
1.9908259040938e-08
|
943
|
89
|
22
|
0.247191011235955
|
0.0233297985153765
|
|
GO:BP
|
GO:0060548
|
negative regulation of cell death
|
2.01706303138448e-08
|
1038
|
89
|
23
|
0.258426966292135
|
0.0221579961464355
|
|
GO:BP
|
GO:0050896
|
response to stimulus
|
2.10692912153179e-08
|
9407
|
92
|
76
|
0.826086956521739
|
0.00807909003933241
|
|
GO:BP
|
GO:0002521
|
leukocyte differentiation
|
2.56820514184532e-08
|
550
|
123
|
20
|
0.16260162601626
|
0.0363636363636364
|
|
GO:BP
|
GO:0031399
|
regulation of protein modification process
|
3.31924184572468e-08
|
1671
|
96
|
30
|
0.3125
|
0.0179533213644524
|
|
GO:BP
|
GO:0048869
|
cellular developmental process
|
3.4387574502829e-08
|
4428
|
101
|
53
|
0.524752475247525
|
0.0119692863595303
|
|
GO:BP
|
GO:0009968
|
negative regulation of signal transduction
|
3.50643106529853e-08
|
1364
|
96
|
27
|
0.28125
|
0.0197947214076246
|
|
GO:BP
|
GO:0009888
|
tissue development
|
3.82060562572854e-08
|
2107
|
116
|
38
|
0.327586206896552
|
0.0180351210251542
|
|
GO:BP
|
GO:1901698
|
response to nitrogen compound
|
4.04582147450259e-08
|
1172
|
76
|
22
|
0.289473684210526
|
0.0187713310580205
|
|
GO:BP
|
GO:0009058
|
biosynthetic process
|
4.64918864542913e-08
|
6340
|
101
|
65
|
0.643564356435644
|
0.0102523659305994
|
|
GO:BP
|
GO:0023052
|
signaling
|
4.84451128886148e-08
|
6810
|
92
|
63
|
0.684782608695652
|
0.0092511013215859
|
|
GO:BP
|
GO:0044237
|
cellular metabolic process
|
5.37211770062543e-08
|
10871
|
101
|
88
|
0.871287128712871
|
0.00809493146904609
|
|
GO:BP
|
GO:0061014
|
positive regulation of mRNA catabolic process
|
5.56220316818478e-08
|
52
|
42
|
6
|
0.142857142857143
|
0.115384615384615
|
|
GO:BP
|
GO:0010243
|
response to organonitrogen compound
|
5.88501720083483e-08
|
1085
|
76
|
21
|
0.276315789473684
|
0.0193548387096774
|
|
GO:BP
|
GO:0031331
|
positive regulation of cellular catabolic process
|
7.99955818158873e-08
|
393
|
46
|
11
|
0.239130434782609
|
0.0279898218829517
|
|
GO:BP
|
GO:0090304
|
nucleic acid metabolic process
|
8.40182856032609e-08
|
5226
|
95
|
55
|
0.578947368421053
|
0.0105243015690777
|
|
GO:BP
|
GO:0051338
|
regulation of transferase activity
|
8.42968803803729e-08
|
1035
|
96
|
23
|
0.239583333333333
|
0.0222222222222222
|
|
GO:BP
|
GO:1902105
|
regulation of leukocyte differentiation
|
1.03698984942108e-07
|
288
|
96
|
13
|
0.135416666666667
|
0.0451388888888889
|
|
GO:BP
|
GO:0045444
|
fat cell differentiation
|
1.03698984942108e-07
|
229
|
120
|
13
|
0.108333333333333
|
0.0567685589519651
|
|
GO:BP
|
GO:0034097
|
response to cytokine
|
1.03698984942108e-07
|
1226
|
58
|
19
|
0.327586206896552
|
0.0154975530179445
|
|
GO:BP
|
GO:0051247
|
positive regulation of protein metabolic process
|
1.03698984942108e-07
|
1564
|
52
|
20
|
0.384615384615385
|
0.0127877237851662
|
|
GO:BP
|
GO:0071363
|
cellular response to growth factor stimulus
|
1.06505062535639e-07
|
731
|
34
|
12
|
0.352941176470588
|
0.0164158686730506
|
|
GO:BP
|
GO:0034599
|
cellular response to oxidative stress
|
1.0779314785649e-07
|
316
|
45
|
10
|
0.222222222222222
|
0.0316455696202532
|
|
GO:BP
|
GO:0009725
|
response to hormone
|
1.08354017344433e-07
|
921
|
23
|
11
|
0.478260869565217
|
0.011943539630836
|
|
GO:BP
|
GO:0065009
|
regulation of molecular function
|
1.10495183013823e-07
|
3221
|
97
|
42
|
0.43298969072165
|
0.0130394287488358
|
|
GO:BP
|
GO:0045859
|
regulation of protein kinase activity
|
1.16889785044049e-07
|
809
|
113
|
22
|
0.194690265486726
|
0.0271940667490729
|
|
GO:BP
|
GO:0044249
|
cellular biosynthetic process
|
1.18767524288918e-07
|
6154
|
101
|
63
|
0.623762376237624
|
0.0102372440688983
|
|
GO:BP
|
GO:1901652
|
response to peptide
|
1.22550683949132e-07
|
551
|
122
|
19
|
0.155737704918033
|
0.0344827586206897
|
|
GO:BP
|
GO:0050789
|
regulation of biological process
|
1.27050275934509e-07
|
12091
|
92
|
85
|
0.923913043478261
|
0.00703002233065917
|
|
GO:BP
|
GO:2000026
|
regulation of multicellular organismal development
|
1.45039223067577e-07
|
1447
|
123
|
31
|
0.252032520325203
|
0.0214236351071182
|
|
GO:BP
|
GO:1900153
|
positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay
|
1.49335770835494e-07
|
15
|
28
|
4
|
0.142857142857143
|
0.266666666666667
|
|
GO:BP
|
GO:0071345
|
cellular response to cytokine stimulus
|
1.83042489968897e-07
|
1132
|
58
|
18
|
0.310344827586207
|
0.0159010600706714
|
|
GO:BP
|
GO:0032496
|
response to lipopolysaccharide
|
2.01927170901383e-07
|
338
|
58
|
11
|
0.189655172413793
|
0.0325443786982249
|
|
GO:BP
|
GO:1901576
|
organic substance biosynthetic process
|
2.21133388653155e-07
|
6248
|
101
|
63
|
0.623762376237624
|
0.0100832266325224
|
|
GO:BP
|
GO:0043549
|
regulation of kinase activity
|
2.28926805010183e-07
|
921
|
104
|
22
|
0.211538461538462
|
0.0238870792616721
|
|
GO:BP
|
GO:0032270
|
positive regulation of cellular protein metabolic process
|
2.41869663452341e-07
|
1477
|
52
|
19
|
0.365384615384615
|
0.012863913337847
|
|
GO:BP
|
GO:1900151
|
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay
|
2.49639231202634e-07
|
17
|
28
|
4
|
0.142857142857143
|
0.235294117647059
|
|
GO:BP
|
GO:0080135
|
regulation of cellular response to stress
|
2.74195203822089e-07
|
725
|
89
|
18
|
0.202247191011236
|
0.0248275862068966
|
|
GO:BP
|
GO:0009653
|
anatomical structure morphogenesis
|
2.9911380382957e-07
|
2797
|
121
|
44
|
0.363636363636364
|
0.0157311405076868
|
|
GO:BP
|
GO:0042127
|
regulation of cell population proliferation
|
3.20640803701893e-07
|
1774
|
41
|
18
|
0.439024390243902
|
0.0101465614430665
|
|
GO:BP
|
GO:0009896
|
positive regulation of catabolic process
|
3.73004055748456e-07
|
464
|
46
|
11
|
0.239130434782609
|
0.0237068965517241
|
|
GO:BP
|
GO:0002237
|
response to molecule of bacterial origin
|
3.75619959191685e-07
|
361
|
58
|
11
|
0.189655172413793
|
0.0304709141274238
|
|
GO:BP
|
GO:0051239
|
regulation of multicellular organismal process
|
3.86254841274591e-07
|
2861
|
100
|
39
|
0.39
|
0.0136315973435862
|
|
GO:BP
|
GO:0071243
|
cellular response to arsenic-containing substance
|
4.37432791823697e-07
|
19
|
83
|
5
|
0.0602409638554217
|
0.263157894736842
|
|
GO:BP
|
GO:0001932
|
regulation of protein phosphorylation
|
4.37432791823697e-07
|
1240
|
96
|
24
|
0.25
|
0.0193548387096774
|
|
GO:BP
|
GO:0051384
|
response to glucocorticoid
|
4.88652517210642e-07
|
146
|
23
|
6
|
0.260869565217391
|
0.0410958904109589
|
|
GO:BP
|
GO:0007519
|
skeletal muscle tissue development
|
4.88652517210642e-07
|
160
|
11
|
5
|
0.454545454545455
|
0.03125
|
|
GO:BP
|
GO:0051174
|
regulation of phosphorus metabolic process
|
5.73509400418561e-07
|
1645
|
104
|
29
|
0.278846153846154
|
0.017629179331307
|
|
GO:BP
|
GO:0019220
|
regulation of phosphate metabolic process
|
5.73509400418561e-07
|
1645
|
104
|
29
|
0.278846153846154
|
0.017629179331307
|
|
GO:BP
|
GO:0007165
|
signal transduction
|
5.82045639847389e-07
|
6309
|
58
|
41
|
0.706896551724138
|
0.00649865271833888
|
|
GO:BP
|
GO:0045597
|
positive regulation of cell differentiation
|
5.88015973910132e-07
|
879
|
96
|
20
|
0.208333333333333
|
0.0227531285551763
|
|
GO:BP
|
GO:0050790
|
regulation of catalytic activity
|
6.1856384523578e-07
|
2545
|
104
|
37
|
0.355769230769231
|
0.0145383104125737
|
|
GO:BP
|
GO:0006807
|
nitrogen compound metabolic process
|
6.24655907491477e-07
|
10078
|
96
|
79
|
0.822916666666667
|
0.00783885691605477
|
|
GO:BP
|
GO:0060538
|
skeletal muscle organ development
|
6.35043083464231e-07
|
170
|
11
|
5
|
0.454545454545455
|
0.0294117647058824
|
|
GO:BP
|
GO:0008152
|
metabolic process
|
6.56707220359574e-07
|
11827
|
92
|
83
|
0.902173913043478
|
0.00701784053437051
|
|
GO:BP
|
GO:0009612
|
response to mechanical stimulus
|
6.61676812674291e-07
|
218
|
78
|
10
|
0.128205128205128
|
0.0458715596330275
|
|
GO:BP
|
GO:0009605
|
response to external stimulus
|
8.60731977075014e-07
|
2944
|
78
|
33
|
0.423076923076923
|
0.0112092391304348
|
|
GO:BP
|
GO:0031960
|
response to corticosteroid
|
9.4803855819252e-07
|
165
|
23
|
6
|
0.260869565217391
|
0.0363636363636364
|
|
GO:BP
|
GO:1903313
|
positive regulation of mRNA metabolic process
|
9.58048759418078e-07
|
87
|
42
|
6
|
0.142857142857143
|
0.0689655172413793
|
|
GO:BP
|
GO:0071495
|
cellular response to endogenous stimulus
|
1.05680240330767e-06
|
1422
|
53
|
18
|
0.339622641509434
|
0.0126582278481013
|
|
GO:BP
|
GO:0006469
|
negative regulation of protein kinase activity
|
1.11090025090375e-06
|
251
|
113
|
12
|
0.106194690265487
|
0.047808764940239
|
|
GO:BP
|
GO:0016310
|
phosphorylation
|
1.11091764173343e-06
|
2138
|
114
|
35
|
0.307017543859649
|
0.0163704396632367
|
|
GO:BP
|
GO:0061158
|
3'-UTR-mediated mRNA destabilization
|
1.1571227684445e-06
|
17
|
42
|
4
|
0.0952380952380952
|
0.235294117647059
|
|
GO:BP
|
GO:0044092
|
negative regulation of molecular function
|
1.31680991365157e-06
|
1219
|
96
|
23
|
0.239583333333333
|
0.0188679245283019
|
|
GO:BP
|
GO:0051348
|
negative regulation of transferase activity
|
1.36541581558209e-06
|
310
|
113
|
13
|
0.115044247787611
|
0.0419354838709677
|
|
GO:BP
|
GO:0042325
|
regulation of phosphorylation
|
1.36541581558209e-06
|
1431
|
96
|
25
|
0.260416666666667
|
0.0174703004891684
|
|
GO:BP
|
GO:0010942
|
positive regulation of cell death
|
1.38807115159871e-06
|
684
|
17
|
8
|
0.470588235294118
|
0.0116959064327485
|
|
GO:BP
|
GO:0051591
|
response to cAMP
|
1.46094230388785e-06
|
97
|
21
|
5
|
0.238095238095238
|
0.0515463917525773
|
|
GO:BP
|
GO:0001933
|
negative regulation of protein phosphorylation
|
1.60677732117286e-06
|
373
|
113
|
14
|
0.123893805309735
|
0.0375335120643432
|
|
GO:BP
|
GO:0044238
|
primary metabolic process
|
1.60677732117286e-06
|
10584
|
101
|
84
|
0.831683168316832
|
0.00793650793650794
|
|
GO:BP
|
GO:0038066
|
p38MAPK cascade
|
1.73695395839809e-06
|
52
|
78
|
6
|
0.0769230769230769
|
0.115384615384615
|
|
GO:BP
|
GO:0031400
|
negative regulation of protein modification process
|
2.03659283372124e-06
|
578
|
113
|
17
|
0.150442477876106
|
0.0294117647058824
|
|
GO:BP
|
GO:1902531
|
regulation of intracellular signal transduction
|
2.2273293895e-06
|
1842
|
89
|
27
|
0.303370786516854
|
0.0146579804560261
|
|
GO:BP
|
GO:0001944
|
vasculature development
|
2.23676449499525e-06
|
830
|
81
|
17
|
0.209876543209877
|
0.0204819277108434
|
|
GO:BP
|
GO:0006139
|
nucleobase-containing compound metabolic process
|
2.23770185231918e-06
|
5746
|
95
|
55
|
0.578947368421053
|
0.00957187608771319
|
|
GO:BP
|
GO:0000288
|
nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay
|
2.33987550454961e-06
|
79
|
28
|
5
|
0.178571428571429
|
0.0632911392405063
|
|
GO:BP
|
GO:0065007
|
biological regulation
|
2.36084860527174e-06
|
12809
|
101
|
93
|
0.920792079207921
|
0.00726051994691233
|
|
GO:BP
|
GO:0051094
|
positive regulation of developmental process
|
2.49076974808869e-06
|
1341
|
53
|
17
|
0.320754716981132
|
0.0126771066368382
|
|
GO:BP
|
GO:0008283
|
cell population proliferation
|
2.5038645958256e-06
|
2054
|
41
|
18
|
0.439024390243902
|
0.00876338851022395
|
|
GO:BP
|
GO:1902895
|
positive regulation of pri-miRNA transcription by RNA polymerase II
|
2.63397015898694e-06
|
40
|
22
|
4
|
0.181818181818182
|
0.1
|
|
GO:BP
|
GO:0031329
|
regulation of cellular catabolic process
|
2.63878832090508e-06
|
866
|
46
|
13
|
0.282608695652174
|
0.0150115473441109
|
|
GO:BP
|
GO:0033673
|
negative regulation of kinase activity
|
2.68604326550628e-06
|
275
|
113
|
12
|
0.106194690265487
|
0.0436363636363636
|
|
GO:BP
|
GO:0043085
|
positive regulation of catalytic activity
|
3.06008971616629e-06
|
1418
|
101
|
25
|
0.247524752475248
|
0.0176304654442877
|
|
GO:BP
|
GO:0044267
|
cellular protein metabolic process
|
3.06024591741491e-06
|
5229
|
96
|
52
|
0.541666666666667
|
0.00994454006502199
|
|
GO:BP
|
GO:0048545
|
response to steroid hormone
|
3.09189796942889e-06
|
349
|
23
|
7
|
0.304347826086957
|
0.0200573065902579
|
|
GO:BP
|
GO:0071248
|
cellular response to metal ion
|
3.18440763607504e-06
|
192
|
6
|
4
|
0.666666666666667
|
0.0208333333333333
|
|
GO:BP
|
GO:0030217
|
T cell differentiation
|
3.651563385445e-06
|
260
|
123
|
12
|
0.0975609756097561
|
0.0461538461538462
|
|
GO:BP
|
GO:0045639
|
positive regulation of myeloid cell differentiation
|
3.66415111183136e-06
|
103
|
24
|
5
|
0.208333333333333
|
0.0485436893203883
|
|
GO:BP
|
GO:0007623
|
circadian rhythm
|
3.66415111183136e-06
|
219
|
119
|
11
|
0.092436974789916
|
0.0502283105022831
|
|
GO:BP
|
GO:0010035
|
response to inorganic substance
|
3.66415111183136e-06
|
575
|
22
|
8
|
0.363636363636364
|
0.0139130434782609
|
|
GO:BP
|
GO:0051093
|
negative regulation of developmental process
|
3.74284306985362e-06
|
977
|
116
|
22
|
0.189655172413793
|
0.022517911975435
|
|
GO:BP
|
GO:1901360
|
organic cyclic compound metabolic process
|
3.88119647569218e-06
|
6201
|
46
|
33
|
0.717391304347826
|
0.00532172230285438
|
|
GO:BP
|
GO:0000165
|
MAPK cascade
|
4.45130048310336e-06
|
935
|
85
|
18
|
0.211764705882353
|
0.0192513368983957
|
|
GO:BP
|
GO:0007049
|
cell cycle
|
4.5312096326899e-06
|
1902
|
95
|
28
|
0.294736842105263
|
0.0147213459516299
|
|
GO:BP
|
GO:0000302
|
response to reactive oxygen species
|
5.05025488337103e-06
|
237
|
51
|
8
|
0.156862745098039
|
0.0337552742616034
|
|
GO:BP
|
GO:0046483
|
heterocycle metabolic process
|
5.10115891645659e-06
|
5904
|
46
|
32
|
0.695652173913043
|
0.00542005420054201
|
|
GO:BP
|
GO:0043618
|
regulation of transcription from RNA polymerase II promoter in response to stress
|
5.18211775531342e-06
|
111
|
45
|
6
|
0.133333333333333
|
0.0540540540540541
|
|
GO:BP
|
GO:0071241
|
cellular response to inorganic substance
|
5.18938417072626e-06
|
220
|
6
|
4
|
0.666666666666667
|
0.0181818181818182
|
|
GO:BP
|
GO:0048584
|
positive regulation of response to stimulus
|
5.28839411121881e-06
|
2344
|
54
|
22
|
0.407407407407407
|
0.00938566552901024
|
|
GO:BP
|
GO:0001568
|
blood vessel development
|
5.2971785236657e-06
|
792
|
53
|
13
|
0.245283018867925
|
0.0164141414141414
|
|
GO:BP
|
GO:0007166
|
cell surface receptor signaling pathway
|
5.31063589451277e-06
|
3210
|
92
|
37
|
0.402173913043478
|
0.0115264797507788
|
|
GO:BP
|
GO:0046685
|
response to arsenic-containing substance
|
5.33959504061468e-06
|
32
|
83
|
5
|
0.0602409638554217
|
0.15625
|
|
GO:BP
|
GO:0042326
|
negative regulation of phosphorylation
|
5.78137648730374e-06
|
420
|
113
|
14
|
0.123893805309735
|
0.0333333333333333
|
|
GO:BP
|
GO:1902893
|
regulation of pri-miRNA transcription by RNA polymerase II
|
5.86656213875054e-06
|
50
|
22
|
4
|
0.181818181818182
|
0.08
|
|
GO:BP
|
GO:0006725
|
cellular aromatic compound metabolic process
|
6.12672428143027e-06
|
5956
|
46
|
32
|
0.695652173913043
|
0.00537273337810611
|
|
GO:BP
|
GO:0046683
|
response to organophosphorus
|
6.15045793971234e-06
|
137
|
9
|
4
|
0.444444444444444
|
0.0291970802919708
|
|
GO:BP
|
GO:0061614
|
pri-miRNA transcription by RNA polymerase II
|
6.27520914208329e-06
|
51
|
22
|
4
|
0.181818181818182
|
0.0784313725490196
|
|
GO:BP
|
GO:0051592
|
response to calcium ion
|
6.34072228423825e-06
|
151
|
19
|
5
|
0.263157894736842
|
0.033112582781457
|
|
GO:BP
|
GO:0010038
|
response to metal ion
|
6.54350968828789e-06
|
371
|
9
|
5
|
0.555555555555556
|
0.0134770889487871
|
|
GO:BP
|
GO:0043620
|
regulation of DNA-templated transcription in response to stress
|
6.67186544749444e-06
|
117
|
45
|
6
|
0.133333333333333
|
0.0512820512820513
|
|
GO:BP
|
GO:0072359
|
circulatory system development
|
6.91813484135722e-06
|
1194
|
100
|
22
|
0.22
|
0.0184254606365159
|
|
GO:BP
|
GO:0071704
|
organic substance metabolic process
|
7.01537405597828e-06
|
11326
|
101
|
86
|
0.851485148514851
|
0.00759314850785803
|
|
GO:BP
|
GO:0045596
|
negative regulation of cell differentiation
|
7.03056265375811e-06
|
697
|
116
|
18
|
0.155172413793103
|
0.0258249641319943
|
|
GO:BP
|
GO:0061061
|
muscle structure development
|
7.16773015846419e-06
|
645
|
100
|
16
|
0.16
|
0.0248062015503876
|
|
GO:BP
|
GO:0051090
|
regulation of DNA-binding transcription factor activity
|
7.23930738822447e-06
|
456
|
51
|
10
|
0.196078431372549
|
0.0219298245614035
|
|
GO:BP
|
GO:0045786
|
negative regulation of cell cycle
|
7.25296894978372e-06
|
647
|
88
|
15
|
0.170454545454545
|
0.0231839258114374
|
|
GO:BP
|
GO:0061013
|
regulation of mRNA catabolic process
|
8.19192200019941e-06
|
213
|
42
|
7
|
0.166666666666667
|
0.0328638497652582
|
|
GO:BP
|
GO:0043065
|
positive regulation of apoptotic process
|
8.39069575017952e-06
|
604
|
95
|
15
|
0.157894736842105
|
0.0248344370860927
|
|
GO:BP
|
GO:0051403
|
stress-activated MAPK cascade
|
8.69984188357549e-06
|
272
|
85
|
10
|
0.117647058823529
|
0.0367647058823529
|
|
GO:BP
|
GO:0002763
|
positive regulation of myeloid leukocyte differentiation
|
8.76375428416556e-06
|
59
|
21
|
4
|
0.19047619047619
|
0.0677966101694915
|
|
GO:BP
|
GO:0014074
|
response to purine-containing compound
|
8.76375428416556e-06
|
152
|
9
|
4
|
0.444444444444444
|
0.0263157894736842
|
|
GO:BP
|
GO:0045637
|
regulation of myeloid cell differentiation
|
8.93015307712441e-06
|
263
|
35
|
7
|
0.2
|
0.026615969581749
|
|
GO:BP
|
GO:1901653
|
cellular response to peptide
|
9.68465852182624e-06
|
408
|
122
|
14
|
0.114754098360656
|
0.0343137254901961
|
|
GO:BP
|
GO:0044093
|
positive regulation of molecular function
|
1.08147022475603e-05
|
1771
|
118
|
30
|
0.254237288135593
|
0.0169395821569735
|
|
GO:BP
|
GO:0048514
|
blood vessel morphogenesis
|
1.09401196882254e-05
|
711
|
53
|
12
|
0.226415094339623
|
0.0168776371308017
|
|
GO:BP
|
GO:0043068
|
positive regulation of programmed cell death
|
1.15042259035364e-05
|
621
|
95
|
15
|
0.157894736842105
|
0.0241545893719807
|
|
GO:BP
|
GO:0006464
|
cellular protein modification process
|
1.17117408221003e-05
|
4041
|
96
|
43
|
0.447916666666667
|
0.0106409304627567
|
|
GO:BP
|
GO:0071496
|
cellular response to external stimulus
|
1.17117408221003e-05
|
322
|
113
|
12
|
0.106194690265487
|
0.0372670807453416
|
|
GO:BP
|
GO:0036211
|
protein modification process
|
1.17117408221003e-05
|
4041
|
96
|
43
|
0.447916666666667
|
0.0106409304627567
|
|
GO:BP
|
GO:0048511
|
rhythmic process
|
1.23488981678545e-05
|
307
|
119
|
12
|
0.100840336134454
|
0.0390879478827362
|
|
GO:BP
|
GO:0007517
|
muscle organ development
|
1.24013764385452e-05
|
333
|
11
|
5
|
0.454545454545455
|
0.015015015015015
|
|
GO:BP
|
GO:0030099
|
myeloid cell differentiation
|
1.26336555447295e-05
|
428
|
45
|
9
|
0.2
|
0.0210280373831776
|
|
GO:BP
|
GO:0060213
|
positive regulation of nuclear-transcribed mRNA poly(A) tail shortening
|
1.34016719159826e-05
|
13
|
28
|
3
|
0.107142857142857
|
0.230769230769231
|
|
GO:BP
|
GO:0071277
|
cellular response to calcium ion
|
1.36241041635767e-05
|
84
|
5
|
3
|
0.6
|
0.0357142857142857
|
|
GO:BP
|
GO:0031098
|
stress-activated protein kinase signaling cascade
|
1.37055596014928e-05
|
288
|
85
|
10
|
0.117647058823529
|
0.0347222222222222
|
|
GO:BP
|
GO:0061157
|
mRNA destabilization
|
1.3757016729346e-05
|
33
|
42
|
4
|
0.0952380952380952
|
0.121212121212121
|
|
GO:BP
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
1.42627323559396e-05
|
6563
|
46
|
33
|
0.717391304347826
|
0.0050281883285083
|
|
GO:BP
|
GO:0009894
|
regulation of catabolic process
|
1.53164464266871e-05
|
1033
|
46
|
13
|
0.282608695652174
|
0.0125847047434656
|
|
GO:BP
|
GO:0071774
|
response to fibroblast growth factor
|
1.67442526657142e-05
|
153
|
23
|
5
|
0.217391304347826
|
0.0326797385620915
|
|
GO:BP
|
GO:0043086
|
negative regulation of catalytic activity
|
1.71068026125856e-05
|
851
|
113
|
19
|
0.168141592920354
|
0.0223266745005875
|
|
GO:BP
|
GO:1902107
|
positive regulation of leukocyte differentiation
|
1.71068026125856e-05
|
157
|
123
|
9
|
0.0731707317073171
|
0.0573248407643312
|
|
GO:BP
|
GO:1903708
|
positive regulation of hemopoiesis
|
1.71068026125856e-05
|
157
|
123
|
9
|
0.0731707317073171
|
0.0573248407643312
|
|
GO:BP
|
GO:0051240
|
positive regulation of multicellular organismal process
|
1.71068026125856e-05
|
1455
|
123
|
27
|
0.219512195121951
|
0.0185567010309278
|
|
GO:BP
|
GO:0043412
|
macromolecule modification
|
1.71131406154412e-05
|
4257
|
96
|
44
|
0.458333333333333
|
0.0103359173126615
|
|
GO:BP
|
GO:0097190
|
apoptotic signaling pathway
|
1.73570184695712e-05
|
600
|
89
|
14
|
0.157303370786517
|
0.0233333333333333
|
|
GO:BP
|
GO:1901654
|
response to ketone
|
1.77656152964165e-05
|
204
|
71
|
8
|
0.112676056338028
|
0.0392156862745098
|
|
GO:BP
|
GO:1903131
|
mononuclear cell differentiation
|
1.812152453895e-05
|
430
|
123
|
14
|
0.113821138211382
|
0.0325581395348837
|
|
GO:BP
|
GO:0071356
|
cellular response to tumor necrosis factor
|
1.87363364735628e-05
|
306
|
65
|
9
|
0.138461538461538
|
0.0294117647058824
|
|
GO:BP
|
GO:0050779
|
RNA destabilization
|
1.87373664078404e-05
|
36
|
42
|
4
|
0.0952380952380952
|
0.111111111111111
|
|
GO:BP
|
GO:0045936
|
negative regulation of phosphate metabolic process
|
1.89459225170262e-05
|
543
|
113
|
15
|
0.132743362831858
|
0.0276243093922652
|
|
GO:BP
|
GO:0010563
|
negative regulation of phosphorus metabolic process
|
1.93101843675125e-05
|
544
|
113
|
15
|
0.132743362831858
|
0.0275735294117647
|
|
GO:BP
|
GO:0007346
|
regulation of mitotic cell cycle
|
1.93794519240706e-05
|
619
|
113
|
16
|
0.141592920353982
|
0.0258481421647819
|
|
GO:BP
|
GO:0060211
|
regulation of nuclear-transcribed mRNA poly(A) tail shortening
|
1.96948237047362e-05
|
15
|
28
|
3
|
0.107142857142857
|
0.2
|
|
GO:BP
|
GO:0009991
|
response to extracellular stimulus
|
2.08565636390616e-05
|
512
|
77
|
12
|
0.155844155844156
|
0.0234375
|
|
GO:BP
|
GO:2001233
|
regulation of apoptotic signaling pathway
|
2.13557141212525e-05
|
363
|
89
|
11
|
0.123595505617978
|
0.0303030303030303
|
|
GO:BP
|
GO:0014706
|
striated muscle tissue development
|
2.15877385409133e-05
|
380
|
11
|
5
|
0.454545454545455
|
0.0131578947368421
|
|
GO:BP
|
GO:0030098
|
lymphocyte differentiation
|
2.20407003409762e-05
|
376
|
123
|
13
|
0.105691056910569
|
0.0345744680851064
|
|
GO:BP
|
GO:1901216
|
positive regulation of neuron death
|
2.36226472812795e-05
|
98
|
17
|
4
|
0.235294117647059
|
0.0408163265306122
|
|
GO:BP
|
GO:0071396
|
cellular response to lipid
|
2.39897942493871e-05
|
578
|
58
|
11
|
0.189655172413793
|
0.0190311418685121
|
|
GO:BP
|
GO:0006402
|
mRNA catabolic process
|
2.45344309024596e-05
|
384
|
29
|
7
|
0.241379310344828
|
0.0182291666666667
|
|
GO:BP
|
GO:0060537
|
muscle tissue development
|
2.69449958049918e-05
|
399
|
11
|
5
|
0.454545454545455
|
0.012531328320802
|
|
GO:BP
|
GO:0019538
|
protein metabolic process
|
2.88889274206676e-05
|
5808
|
96
|
53
|
0.552083333333333
|
0.00912534435261708
|
|
GO:BP
|
GO:0043434
|
response to peptide hormone
|
2.93631343980324e-05
|
462
|
120
|
14
|
0.116666666666667
|
0.0303030303030303
|
|
GO:BP
|
GO:0006796
|
phosphate-containing compound metabolic process
|
3.12075688438836e-05
|
3087
|
114
|
40
|
0.350877192982456
|
0.0129575639779721
|
|
GO:BP
|
GO:0035239
|
tube morphogenesis
|
3.14861034537872e-05
|
950
|
53
|
13
|
0.245283018867925
|
0.0136842105263158
|
|
GO:BP
|
GO:0034612
|
response to tumor necrosis factor
|
3.34567024270955e-05
|
331
|
65
|
9
|
0.138461538461538
|
0.027190332326284
|
|
GO:BP
|
GO:0071376
|
cellular response to corticotropin-releasing hormone stimulus
|
3.5242679840416e-05
|
5
|
120
|
3
|
0.025
|
0.6
|
|
GO:BP
|
GO:0043435
|
response to corticotropin-releasing hormone
|
3.5242679840416e-05
|
5
|
120
|
3
|
0.025
|
0.6
|
|
GO:BP
|
GO:0071385
|
cellular response to glucocorticoid stimulus
|
3.58292321131072e-05
|
57
|
71
|
5
|
0.0704225352112676
|
0.087719298245614
|
|
GO:BP
|
GO:0071900
|
regulation of protein serine/threonine kinase activity
|
3.73150365042129e-05
|
503
|
113
|
14
|
0.123893805309735
|
0.0278330019880716
|
|
GO:BP
|
GO:0006793
|
phosphorus metabolic process
|
3.8053455037979e-05
|
3114
|
114
|
40
|
0.350877192982456
|
0.0128452151573539
|
|
GO:BP
|
GO:0044706
|
multi-multicellular organism process
|
3.99333612991074e-05
|
227
|
72
|
8
|
0.111111111111111
|
0.0352422907488987
|
|
GO:BP
|
GO:0031668
|
cellular response to extracellular stimulus
|
4.29575312149325e-05
|
248
|
113
|
10
|
0.0884955752212389
|
0.0403225806451613
|
|
GO:BP
|
GO:0006401
|
RNA catabolic process
|
4.49870847436419e-05
|
424
|
29
|
7
|
0.241379310344828
|
0.0165094339622642
|
|
GO:BP
|
GO:0002573
|
myeloid leukocyte differentiation
|
4.58458350840342e-05
|
213
|
21
|
5
|
0.238095238095238
|
0.0234741784037559
|
|
GO:BP
|
GO:0045619
|
regulation of lymphocyte differentiation
|
4.64828129925321e-05
|
180
|
123
|
9
|
0.0731707317073171
|
0.05
|
|
GO:BP
|
GO:0035295
|
tube development
|
4.74291240697673e-05
|
1146
|
92
|
19
|
0.206521739130435
|
0.0165794066317627
|
|
GO:BP
|
GO:0007178
|
transmembrane receptor protein serine/threonine kinase signaling pathway
|
5.52265158833635e-05
|
369
|
34
|
7
|
0.205882352941176
|
0.018970189701897
|
|
GO:BP
|
GO:0071384
|
cellular response to corticosteroid stimulus
|
5.64845104971196e-05
|
63
|
71
|
5
|
0.0704225352112676
|
0.0793650793650794
|
|
GO:BP
|
GO:0042594
|
response to starvation
|
5.83558329161428e-05
|
202
|
113
|
9
|
0.079646017699115
|
0.0445544554455446
|
|
GO:BP
|
GO:0045604
|
regulation of epidermal cell differentiation
|
6.01345338948471e-05
|
63
|
72
|
5
|
0.0694444444444444
|
0.0793650793650794
|
|
GO:BP
|
GO:0032570
|
response to progesterone
|
6.05830531017141e-05
|
45
|
14
|
3
|
0.214285714285714
|
0.0666666666666667
|
|
GO:BP
|
GO:0009314
|
response to radiation
|
6.35217970003214e-05
|
461
|
19
|
6
|
0.315789473684211
|
0.0130151843817787
|
|
GO:BP
|
GO:0032870
|
cellular response to hormone stimulus
|
6.48988591293803e-05
|
645
|
120
|
16
|
0.133333333333333
|
0.0248062015503876
|
|
GO:BP
|
GO:0090084
|
negative regulation of inclusion body assembly
|
7.24408168942189e-05
|
11
|
62
|
3
|
0.0483870967741935
|
0.272727272727273
|
|
GO:BP
|
GO:0016477
|
cell migration
|
7.35407702695973e-05
|
1649
|
62
|
18
|
0.290322580645161
|
0.0109157064887811
|
|
GO:BP
|
GO:0006986
|
response to unfolded protein
|
7.57696608030772e-05
|
184
|
97
|
8
|
0.0824742268041237
|
0.0434782608695652
|
|
GO:BP
|
GO:0051674
|
localization of cell
|
7.57720690419117e-05
|
1828
|
62
|
19
|
0.306451612903226
|
0.0103938730853392
|
|
GO:BP
|
GO:0048870
|
cell motility
|
7.57720690419117e-05
|
1828
|
62
|
19
|
0.306451612903226
|
0.0103938730853392
|
|
GO:BP
|
GO:2000379
|
positive regulation of reactive oxygen species metabolic process
|
7.73677799150465e-05
|
107
|
78
|
6
|
0.0769230769230769
|
0.0560747663551402
|
|
GO:BP
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
7.74259530328807e-05
|
1202
|
116
|
22
|
0.189655172413793
|
0.0183028286189684
|
|
GO:BP
|
GO:0042542
|
response to hydrogen peroxide
|
8.06635492486581e-05
|
150
|
120
|
8
|
0.0666666666666667
|
0.0533333333333333
|
|
GO:BP
|
GO:0071560
|
cellular response to transforming growth factor beta stimulus
|
8.06783430326062e-05
|
256
|
51
|
7
|
0.137254901960784
|
0.02734375
|
|
GO:BP
|
GO:0009267
|
cellular response to starvation
|
8.25586574307553e-05
|
160
|
113
|
8
|
0.0707964601769911
|
0.05
|
|
GO:BP
|
GO:0007565
|
female pregnancy
|
9.18761161462006e-05
|
198
|
26
|
5
|
0.192307692307692
|
0.0252525252525253
|
|
GO:BP
|
GO:1902532
|
negative regulation of intracellular signal transduction
|
9.22979630498851e-05
|
549
|
113
|
14
|
0.123893805309735
|
0.0255009107468124
|
|
GO:BP
|
GO:0071559
|
response to transforming growth factor beta
|
9.26658700079495e-05
|
262
|
51
|
7
|
0.137254901960784
|
0.0267175572519084
|
|
GO:BP
|
GO:0045682
|
regulation of epidermis development
|
9.59034440775127e-05
|
70
|
72
|
5
|
0.0694444444444444
|
0.0714285714285714
|
|
GO:BP
|
GO:1901214
|
regulation of neuron death
|
0.000100560673495138
|
323
|
17
|
5
|
0.294117647058824
|
0.0154798761609907
|
|
GO:BP
|
GO:0051241
|
negative regulation of multicellular organismal process
|
0.000100560673495138
|
1160
|
96
|
19
|
0.197916666666667
|
0.0163793103448276
|
|
GO:BP
|
GO:0097191
|
extrinsic apoptotic signaling pathway
|
0.000100560673495138
|
223
|
84
|
8
|
0.0952380952380952
|
0.0358744394618834
|
|
GO:BP
|
GO:0001666
|
response to hypoxia
|
0.000100560673495138
|
367
|
52
|
8
|
0.153846153846154
|
0.0217983651226158
|
|
GO:BP
|
GO:2001236
|
regulation of extrinsic apoptotic signaling pathway
|
0.000100560673495138
|
159
|
84
|
7
|
0.0833333333333333
|
0.0440251572327044
|
|
GO:BP
|
GO:0000956
|
nuclear-transcribed mRNA catabolic process
|
0.000102845138351391
|
214
|
42
|
6
|
0.142857142857143
|
0.0280373831775701
|
|
GO:BP
|
GO:0036499
|
PERK-mediated unfolded protein response
|
0.000104689187750415
|
23
|
33
|
3
|
0.0909090909090909
|
0.130434782608696
|
|
GO:BP
|
GO:0071417
|
cellular response to organonitrogen compound
|
0.000112135882296824
|
666
|
122
|
16
|
0.131147540983607
|
0.024024024024024
|
|
GO:BP
|
GO:0002761
|
regulation of myeloid leukocyte differentiation
|
0.00012074695871442
|
123
|
21
|
4
|
0.19047619047619
|
0.032520325203252
|
|
GO:BP
|
GO:0045862
|
positive regulation of proteolysis
|
0.00012234555961534
|
378
|
52
|
8
|
0.153846153846154
|
0.0211640211640212
|
|
GO:BP
|
GO:0017148
|
negative regulation of translation
|
0.000127674975598994
|
223
|
42
|
6
|
0.142857142857143
|
0.0269058295964126
|
|
GO:BP
|
GO:0001701
|
in utero embryonic development
|
0.000130254021631973
|
365
|
89
|
10
|
0.112359550561798
|
0.0273972602739726
|
|
GO:BP
|
GO:0036293
|
response to decreased oxygen levels
|
0.000130846979445856
|
382
|
52
|
8
|
0.153846153846154
|
0.0209424083769634
|
|
GO:BP
|
GO:0071347
|
cellular response to interleukin-1
|
0.000138062467789778
|
185
|
51
|
6
|
0.117647058823529
|
0.0324324324324324
|
|
GO:BP
|
GO:0009266
|
response to temperature stimulus
|
0.00013906128345166
|
228
|
113
|
9
|
0.079646017699115
|
0.0394736842105263
|
|
GO:BP
|
GO:0043405
|
regulation of MAP kinase activity
|
0.000148016228408371
|
316
|
104
|
10
|
0.0961538461538462
|
0.0316455696202532
|
|
GO:BP
|
GO:0035966
|
response to topologically incorrect protein
|
0.000152319067357794
|
205
|
97
|
8
|
0.0824742268041237
|
0.0390243902439024
|
|
GO:BP
|
GO:1990868
|
response to chemokine
|
0.000153588612536029
|
97
|
58
|
5
|
0.0862068965517241
|
0.0515463917525773
|
|
GO:BP
|
GO:1990869
|
cellular response to chemokine
|
0.000153588612536029
|
97
|
58
|
5
|
0.0862068965517241
|
0.0515463917525773
|
|
GO:BP
|
GO:0040011
|
locomotion
|
0.000153694577392817
|
2028
|
91
|
25
|
0.274725274725275
|
0.01232741617357
|
|
GO:BP
|
GO:0140467
|
integrated stress response signaling
|
0.000164436082734343
|
27
|
33
|
3
|
0.0909090909090909
|
0.111111111111111
|
|
GO:BP
|
GO:1903311
|
regulation of mRNA metabolic process
|
0.000166927267909937
|
356
|
42
|
7
|
0.166666666666667
|
0.0196629213483146
|
|
GO:BP
|
GO:0060395
|
SMAD protein signal transduction
|
0.000170465869748604
|
82
|
34
|
4
|
0.117647058823529
|
0.0487804878048781
|
|
GO:BP
|
GO:0070997
|
neuron death
|
0.000170686102441955
|
365
|
17
|
5
|
0.294117647058824
|
0.0136986301369863
|
|
GO:BP
|
GO:1901991
|
negative regulation of mitotic cell cycle phase transition
|
0.000172089090617798
|
262
|
23
|
5
|
0.217391304347826
|
0.0190839694656489
|
|
GO:BP
|
GO:0045598
|
regulation of fat cell differentiation
|
0.000173596213401232
|
139
|
42
|
5
|
0.119047619047619
|
0.0359712230215827
|
|
GO:BP
|
GO:2001234
|
negative regulation of apoptotic signaling pathway
|
0.000176618714374319
|
236
|
113
|
9
|
0.079646017699115
|
0.038135593220339
|
|
GO:BP
|
GO:0043542
|
endothelial cell migration
|
0.000178503493834163
|
290
|
116
|
10
|
0.0862068965517241
|
0.0344827586206897
|
|
GO:BP
|
GO:0034614
|
cellular response to reactive oxygen species
|
0.000180351178796497
|
170
|
120
|
8
|
0.0666666666666667
|
0.0470588235294118
|
|
GO:BP
|
GO:0070482
|
response to oxygen levels
|
0.000198385440499075
|
408
|
52
|
8
|
0.153846153846154
|
0.0196078431372549
|
|
GO:BP
|
GO:1902106
|
negative regulation of leukocyte differentiation
|
0.00020203757960005
|
105
|
96
|
6
|
0.0625
|
0.0571428571428571
|
|
GO:BP
|
GO:0009617
|
response to bacterium
|
0.000202408671319199
|
743
|
58
|
11
|
0.189655172413793
|
0.0148048452220727
|
|
GO:BP
|
GO:0002682
|
regulation of immune system process
|
0.00021331330649449
|
1617
|
81
|
20
|
0.246913580246914
|
0.0123685837971552
|
|
GO:BP
|
GO:0043433
|
negative regulation of DNA-binding transcription factor activity
|
0.000226345441469757
|
187
|
113
|
8
|
0.0707964601769911
|
0.0427807486631016
|
|
GO:BP
|
GO:1901988
|
negative regulation of cell cycle phase transition
|
0.000232208253596047
|
281
|
23
|
5
|
0.217391304347826
|
0.0177935943060498
|
|
GO:BP
|
GO:0034249
|
negative regulation of cellular amide metabolic process
|
0.00024031056996909
|
253
|
42
|
6
|
0.142857142857143
|
0.0237154150197628
|
|
GO:BP
|
GO:0034655
|
nucleobase-containing compound catabolic process
|
0.000241843922083876
|
567
|
29
|
7
|
0.241379310344828
|
0.0123456790123457
|
|
GO:BP
|
GO:0071364
|
cellular response to epidermal growth factor stimulus
|
0.000243301494049601
|
45
|
23
|
3
|
0.130434782608696
|
0.0666666666666667
|
|
GO:BP
|
GO:1903707
|
negative regulation of hemopoiesis
|
0.000244383643705002
|
109
|
96
|
6
|
0.0625
|
0.055045871559633
|
|
GO:BP
|
GO:0000289
|
nuclear-transcribed mRNA poly(A) tail shortening
|
0.000244972510701985
|
37
|
28
|
3
|
0.107142857142857
|
0.0810810810810811
|
|
GO:BP
|
GO:0019221
|
cytokine-mediated signaling pathway
|
0.000246946678499141
|
810
|
75
|
13
|
0.173333333333333
|
0.0160493827160494
|
|
GO:BP
|
GO:0090083
|
regulation of inclusion body assembly
|
0.000246946678499141
|
17
|
62
|
3
|
0.0483870967741935
|
0.176470588235294
|
|
GO:BP
|
GO:0000278
|
mitotic cell cycle
|
0.000251437550659355
|
1059
|
122
|
20
|
0.163934426229508
|
0.0188857412653447
|
|
GO:BP
|
GO:0010631
|
epithelial cell migration
|
0.000251437550659355
|
370
|
116
|
11
|
0.0948275862068965
|
0.0297297297297297
|
|
GO:BP
|
GO:0009967
|
positive regulation of signal transduction
|
0.000255036747614274
|
1598
|
83
|
20
|
0.240963855421687
|
0.0125156445556946
|
|
GO:BP
|
GO:2000377
|
regulation of reactive oxygen species metabolic process
|
0.000261181378340521
|
203
|
78
|
7
|
0.0897435897435897
|
0.0344827586206897
|
|
GO:BP
|
GO:0090132
|
epithelium migration
|
0.000267756333485167
|
373
|
116
|
11
|
0.0948275862068965
|
0.0294906166219839
|
|
GO:BP
|
GO:1903320
|
regulation of protein modification by small protein conjugation or removal
|
0.000270533358857019
|
243
|
90
|
8
|
0.0888888888888889
|
0.0329218106995885
|
|
GO:BP
|
GO:0042110
|
T cell activation
|
0.000272390889444075
|
489
|
123
|
13
|
0.105691056910569
|
0.0265848670756646
|
|
GO:BP
|
GO:1901699
|
cellular response to nitrogen compound
|
0.00027278389975655
|
724
|
122
|
16
|
0.131147540983607
|
0.0220994475138122
|
|
GO:BP
|
GO:2000045
|
regulation of G1/S transition of mitotic cell cycle
|
0.000281333308911547
|
182
|
88
|
7
|
0.0795454545454545
|
0.0384615384615385
|
|
GO:BP
|
GO:0002376
|
immune system process
|
0.000285761644349545
|
3278
|
58
|
24
|
0.413793103448276
|
0.0073215375228798
|
|
GO:BP
|
GO:0070555
|
response to interleukin-1
|
0.000285761644349545
|
215
|
51
|
6
|
0.117647058823529
|
0.027906976744186
|
|
GO:BP
|
GO:0071456
|
cellular response to hypoxia
|
0.000294846902916417
|
212
|
52
|
6
|
0.115384615384615
|
0.0283018867924528
|
|
GO:BP
|
GO:0045638
|
negative regulation of myeloid cell differentiation
|
0.000297545586891915
|
94
|
35
|
4
|
0.114285714285714
|
0.0425531914893617
|
|
GO:BP
|
GO:0070849
|
response to epidermal growth factor
|
0.000299027093827378
|
49
|
23
|
3
|
0.130434782608696
|
0.0612244897959184
|
|
GO:BP
|
GO:0045646
|
regulation of erythrocyte differentiation
|
0.000299927733884989
|
47
|
24
|
3
|
0.125
|
0.0638297872340425
|
|
GO:BP
|
GO:0090130
|
tissue migration
|
0.000300832979400768
|
379
|
116
|
11
|
0.0948275862068965
|
0.029023746701847
|
|
GO:BP
|
GO:0044265
|
cellular macromolecule catabolic process
|
0.0003026359743604
|
1231
|
52
|
13
|
0.25
|
0.0105605199025183
|
|
GO:BP
|
GO:0060711
|
labyrinthine layer development
|
0.000302788290903857
|
44
|
75
|
4
|
0.0533333333333333
|
0.0909090909090909
|
|
GO:BP
|
GO:0044344
|
cellular response to fibroblast growth factor stimulus
|
0.000304223944073429
|
147
|
23
|
4
|
0.173913043478261
|
0.0272108843537415
|
|
GO:BP
|
GO:0043408
|
regulation of MAPK cascade
|
0.000312714395625384
|
730
|
85
|
13
|
0.152941176470588
|
0.0178082191780822
|
|
GO:BP
|
GO:0010629
|
negative regulation of gene expression
|
0.000319281282298412
|
1460
|
51
|
14
|
0.274509803921569
|
0.00958904109589041
|
|
GO:BP
|
GO:0072593
|
reactive oxygen species metabolic process
|
0.000321219632487075
|
290
|
78
|
8
|
0.102564102564103
|
0.0275862068965517
|
|
GO:BP
|
GO:0046700
|
heterocycle catabolic process
|
0.00032831136595449
|
602
|
29
|
7
|
0.241379310344828
|
0.0116279069767442
|
|
GO:BP
|
GO:0035994
|
response to muscle stretch
|
0.000328739975265448
|
23
|
51
|
3
|
0.0588235294117647
|
0.130434782608696
|
|
GO:BP
|
GO:0031667
|
response to nutrient levels
|
0.000329908486686179
|
482
|
77
|
10
|
0.12987012987013
|
0.020746887966805
|
|
GO:BP
|
GO:0043154
|
negative regulation of cysteine-type endopeptidase activity involved in apoptotic process
|
0.000337563679508657
|
81
|
84
|
5
|
0.0595238095238095
|
0.0617283950617284
|
|
GO:BP
|
GO:0044270
|
cellular nitrogen compound catabolic process
|
0.000346176293527812
|
608
|
29
|
7
|
0.241379310344828
|
0.0115131578947368
|
|
GO:BP
|
GO:2001243
|
negative regulation of intrinsic apoptotic signaling pathway
|
0.000346923347338816
|
100
|
113
|
6
|
0.0530973451327434
|
0.06
|
|
GO:BP
|
GO:0009790
|
embryo development
|
0.000356122431917697
|
1035
|
89
|
16
|
0.179775280898876
|
0.0154589371980676
|
|
GO:BP
|
GO:0045428
|
regulation of nitric oxide biosynthetic process
|
0.000358515793132463
|
68
|
51
|
4
|
0.0784313725490196
|
0.0588235294117647
|
|
GO:BP
|
GO:0048598
|
embryonic morphogenesis
|
0.000358515793132463
|
595
|
120
|
14
|
0.116666666666667
|
0.0235294117647059
|
|
GO:BP
|
GO:0036294
|
cellular response to decreased oxygen levels
|
0.000362355365061712
|
222
|
52
|
6
|
0.115384615384615
|
0.027027027027027
|
|
GO:BP
|
GO:0071222
|
cellular response to lipopolysaccharide
|
0.000378642780314823
|
200
|
58
|
6
|
0.103448275862069
|
0.03
|
|
GO:BP
|
GO:1903364
|
positive regulation of cellular protein catabolic process
|
0.000384582369385535
|
154
|
46
|
5
|
0.108695652173913
|
0.0324675324675325
|
|
GO:BP
|
GO:0071901
|
negative regulation of protein serine/threonine kinase activity
|
0.000384582369385535
|
150
|
113
|
7
|
0.0619469026548673
|
0.0466666666666667
|
|
GO:BP
|
GO:0031401
|
positive regulation of protein modification process
|
0.000394720704000591
|
1074
|
96
|
17
|
0.177083333333333
|
0.015828677839851
|
|
GO:BP
|
GO:0019439
|
aromatic compound catabolic process
|
0.000394720704000591
|
623
|
29
|
7
|
0.241379310344828
|
0.0112359550561798
|
|
GO:BP
|
GO:0080164
|
regulation of nitric oxide metabolic process
|
0.000394974610058959
|
70
|
51
|
4
|
0.0784313725490196
|
0.0571428571428571
|
|
GO:BP
|
GO:0044419
|
biological process involved in interspecies interaction between organisms
|
0.000398394241858056
|
2255
|
58
|
19
|
0.327586206896552
|
0.00842572062084257
|
|
GO:BP
|
GO:1900744
|
regulation of p38MAPK cascade
|
0.000399152053862571
|
46
|
78
|
4
|
0.0512820512820513
|
0.0869565217391304
|
|
GO:BP
|
GO:0030162
|
regulation of proteolysis
|
0.000409300664875825
|
752
|
52
|
10
|
0.192307692307692
|
0.0132978723404255
|
|
GO:BP
|
GO:0043009
|
chordate embryonic development
|
0.000409487858287261
|
616
|
89
|
12
|
0.134831460674157
|
0.0194805194805195
|
|
GO:BP
|
GO:0070371
|
ERK1 and ERK2 cascade
|
0.000431587684574298
|
329
|
23
|
5
|
0.217391304347826
|
0.0151975683890578
|
|
GO:BP
|
GO:0045930
|
negative regulation of mitotic cell cycle
|
0.000455989703911733
|
333
|
23
|
5
|
0.217391304347826
|
0.015015015015015
|
|
GO:BP
|
GO:0001892
|
embryonic placenta development
|
0.000478243704635733
|
83
|
89
|
5
|
0.0561797752808989
|
0.0602409638554217
|
|
GO:BP
|
GO:0001709
|
cell fate determination
|
0.000495856750420832
|
44
|
31
|
3
|
0.0967741935483871
|
0.0681818181818182
|
|
GO:BP
|
GO:0060326
|
cell chemotaxis
|
0.000495856750420832
|
310
|
58
|
7
|
0.120689655172414
|
0.0225806451612903
|
|
GO:BP
|
GO:2000117
|
negative regulation of cysteine-type endopeptidase activity
|
0.00050165125148686
|
89
|
84
|
5
|
0.0595238095238095
|
0.0561797752808989
|
|
GO:BP
|
GO:1901361
|
organic cyclic compound catabolic process
|
0.000504623136719597
|
651
|
29
|
7
|
0.241379310344828
|
0.010752688172043
|
|
GO:BP
|
GO:0007612
|
learning
|
0.000507197301899041
|
146
|
10
|
3
|
0.3
|
0.0205479452054795
|
|
GO:BP
|
GO:0070301
|
cellular response to hydrogen peroxide
|
0.000512030428431176
|
102
|
120
|
6
|
0.05
|
0.0588235294117647
|
|
GO:BP
|
GO:0071219
|
cellular response to molecule of bacterial origin
|
0.000521531695798768
|
214
|
58
|
6
|
0.103448275862069
|
0.0280373831775701
|
|
GO:BP
|
GO:0070423
|
nucleotide-binding oligomerization domain containing signaling pathway
|
0.000523791507236264
|
40
|
35
|
3
|
0.0857142857142857
|
0.075
|
|
GO:BP
|
GO:0009408
|
response to heat
|
0.000523791507236264
|
159
|
113
|
7
|
0.0619469026548673
|
0.0440251572327044
|
|
GO:BP
|
GO:0071453
|
cellular response to oxygen levels
|
0.000523791507236264
|
240
|
52
|
6
|
0.115384615384615
|
0.025
|
|
GO:BP
|
GO:0006417
|
regulation of translation
|
0.000523791507236264
|
462
|
28
|
6
|
0.214285714285714
|
0.012987012987013
|
|
GO:BP
|
GO:0009792
|
embryo development ending in birth or egg hatching
|
0.000523828917158877
|
635
|
89
|
12
|
0.134831460674157
|
0.0188976377952756
|
|
GO:BP
|
GO:1902806
|
regulation of cell cycle G1/S phase transition
|
0.000541170635785627
|
206
|
88
|
7
|
0.0795454545454545
|
0.0339805825242718
|
|
GO:BP
|
GO:0030330
|
DNA damage response, signal transduction by p53 class mediator
|
0.000544097144787092
|
110
|
113
|
6
|
0.0530973451327434
|
0.0545454545454545
|
|
GO:BP
|
GO:1901990
|
regulation of mitotic cell cycle phase transition
|
0.000548175969595083
|
451
|
88
|
10
|
0.113636363636364
|
0.0221729490022173
|
|
GO:BP
|
GO:0060429
|
epithelium development
|
0.000548175969595083
|
1325
|
89
|
18
|
0.202247191011236
|
0.0135849056603774
|
|
GO:BP
|
GO:0035872
|
nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway
|
0.000555241516462037
|
41
|
35
|
3
|
0.0857142857142857
|
0.0731707317073171
|
|
GO:BP
|
GO:0051347
|
positive regulation of transferase activity
|
0.000566442053611579
|
689
|
96
|
13
|
0.135416666666667
|
0.0188679245283019
|
|
GO:BP
|
GO:0030522
|
intracellular receptor signaling pathway
|
0.000566442053611579
|
284
|
45
|
6
|
0.133333333333333
|
0.0211267605633803
|
|
GO:BP
|
GO:0031669
|
cellular response to nutrient levels
|
0.000569603106118886
|
219
|
113
|
8
|
0.0707964601769911
|
0.0365296803652968
|
|
GO:BP
|
GO:1905898
|
positive regulation of response to endoplasmic reticulum stress
|
0.000593954287254762
|
36
|
113
|
4
|
0.0353982300884956
|
0.111111111111111
|
|
GO:BP
|
GO:0070841
|
inclusion body assembly
|
0.000600221250179699
|
24
|
62
|
3
|
0.0483870967741935
|
0.125
|
|
GO:BP
|
GO:0043488
|
regulation of mRNA stability
|
0.000625059469589323
|
191
|
42
|
5
|
0.119047619047619
|
0.0261780104712042
|
|
GO:BP
|
GO:0071383
|
cellular response to steroid hormone stimulus
|
0.000642444712006399
|
210
|
120
|
8
|
0.0666666666666667
|
0.0380952380952381
|
|
GO:BP
|
GO:0032436
|
positive regulation of proteasomal ubiquitin-dependent protein catabolic process
|
0.000643544490402442
|
90
|
46
|
4
|
0.0869565217391304
|
0.0444444444444444
|
|
GO:BP
|
GO:0045580
|
regulation of T cell differentiation
|
0.000650592589615912
|
152
|
123
|
7
|
0.0569105691056911
|
0.0460526315789474
|
|
GO:BP
|
GO:0006809
|
nitric oxide biosynthetic process
|
0.000669954270996219
|
82
|
51
|
4
|
0.0784313725490196
|
0.0487804878048781
|
|
GO:BP
|
GO:0051101
|
regulation of DNA binding
|
0.000678300545624682
|
121
|
35
|
4
|
0.114285714285714
|
0.0330578512396694
|
|
GO:BP
|
GO:2001242
|
regulation of intrinsic apoptotic signaling pathway
|
0.000679977321210532
|
167
|
113
|
7
|
0.0619469026548673
|
0.0419161676646707
|
|
GO:BP
|
GO:0010948
|
negative regulation of cell cycle process
|
0.000704708367042532
|
372
|
23
|
5
|
0.217391304347826
|
0.0134408602150538
|
|
GO:BP
|
GO:0048660
|
regulation of smooth muscle cell proliferation
|
0.000706910231830301
|
174
|
25
|
4
|
0.16
|
0.0229885057471264
|
|
GO:BP
|
GO:0071375
|
cellular response to peptide hormone stimulus
|
0.000706910231830301
|
340
|
120
|
10
|
0.0833333333333333
|
0.0294117647058824
|
|
GO:BP
|
GO:1903426
|
regulation of reactive oxygen species biosynthetic process
|
0.000720662988943911
|
111
|
74
|
5
|
0.0675675675675676
|
0.045045045045045
|
|
GO:BP
|
GO:0098609
|
cell-cell adhesion
|
0.000721262054058694
|
894
|
57
|
11
|
0.192982456140351
|
0.0123042505592841
|
|
GO:BP
|
GO:0048659
|
smooth muscle cell proliferation
|
0.000733981380556817
|
176
|
25
|
4
|
0.16
|
0.0227272727272727
|
|
GO:BP
|
GO:0043487
|
regulation of RNA stability
|
0.000768833682200581
|
201
|
42
|
5
|
0.119047619047619
|
0.0248756218905473
|
|
GO:BP
|
GO:0061469
|
regulation of type B pancreatic cell proliferation
|
0.000769913339891459
|
14
|
120
|
3
|
0.025
|
0.214285714285714
|
|
GO:BP
|
GO:0045937
|
positive regulation of phosphate metabolic process
|
0.00078148991281728
|
1001
|
99
|
16
|
0.161616161616162
|
0.015984015984016
|
|
GO:BP
|
GO:0010562
|
positive regulation of phosphorus metabolic process
|
0.00078148991281728
|
1001
|
99
|
16
|
0.161616161616162
|
0.015984015984016
|
|
GO:BP
|
GO:0070373
|
negative regulation of ERK1 and ERK2 cascade
|
0.000800346716110805
|
81
|
104
|
5
|
0.0480769230769231
|
0.0617283950617284
|
|
GO:BP
|
GO:1903037
|
regulation of leukocyte cell-cell adhesion
|
0.000809731677430045
|
338
|
123
|
10
|
0.0813008130081301
|
0.029585798816568
|
|
GO:BP
|
GO:0046209
|
nitric oxide metabolic process
|
0.000810726138285283
|
87
|
51
|
4
|
0.0784313725490196
|
0.0459770114942529
|
|
GO:BP
|
GO:2001057
|
reactive nitrogen species metabolic process
|
0.000845732785354252
|
88
|
51
|
4
|
0.0784313725490196
|
0.0454545454545455
|
|
GO:BP
|
GO:0071216
|
cellular response to biotic stimulus
|
0.000851319230050595
|
238
|
58
|
6
|
0.103448275862069
|
0.0252100840336134
|
|
GO:BP
|
GO:1903034
|
regulation of response to wounding
|
0.000851319230050595
|
169
|
51
|
5
|
0.0980392156862745
|
0.029585798816568
|
|
GO:BP
|
GO:0043409
|
negative regulation of MAPK cascade
|
0.000865347219583573
|
190
|
104
|
7
|
0.0673076923076923
|
0.0368421052631579
|
|
GO:BP
|
GO:0010647
|
positive regulation of cell communication
|
0.000896512541750575
|
1777
|
83
|
20
|
0.240963855421687
|
0.0112549240292628
|
|
GO:BP
|
GO:1903428
|
positive regulation of reactive oxygen species biosynthetic process
|
0.00091505113554564
|
62
|
74
|
4
|
0.0540540540540541
|
0.0645161290322581
|
|
GO:BP
|
GO:0046632
|
alpha-beta T cell differentiation
|
0.000921683919299648
|
113
|
123
|
6
|
0.0487804878048781
|
0.0530973451327434
|
|
GO:BP
|
GO:0045860
|
positive regulation of protein kinase activity
|
0.000921683919299648
|
534
|
96
|
11
|
0.114583333333333
|
0.0205992509363296
|
|
GO:BP
|
GO:0006984
|
ER-nucleus signaling pathway
|
0.000921683919299648
|
53
|
33
|
3
|
0.0909090909090909
|
0.0566037735849057
|
|
GO:BP
|
GO:0023056
|
positive regulation of signaling
|
0.000921865624765921
|
1782
|
83
|
20
|
0.240963855421687
|
0.0112233445566779
|
|
GO:BP
|
GO:0010628
|
positive regulation of gene expression
|
0.000937287067176429
|
1160
|
62
|
13
|
0.209677419354839
|
0.0112068965517241
|
|
GO:BP
|
GO:1901564
|
organonitrogen compound metabolic process
|
0.000937287067176429
|
6792
|
46
|
30
|
0.652173913043478
|
0.00441696113074205
|
|
GO:BP
|
GO:0000082
|
G1/S transition of mitotic cell cycle
|
0.000959301782667528
|
283
|
122
|
9
|
0.0737704918032787
|
0.0318021201413428
|
|
GO:BP
|
GO:0040012
|
regulation of locomotion
|
0.00096627516584526
|
1104
|
121
|
19
|
0.15702479338843
|
0.0172101449275362
|
|
GO:BP
|
GO:1903047
|
mitotic cell cycle process
|
0.000969287872374772
|
911
|
24
|
7
|
0.291666666666667
|
0.00768386388583974
|
|
GO:BP
|
GO:0045581
|
negative regulation of T cell differentiation
|
0.000976782536686112
|
49
|
96
|
4
|
0.0416666666666667
|
0.0816326530612245
|
|
GO:BP
|
GO:0001934
|
positive regulation of protein phosphorylation
|
0.000988242719037114
|
840
|
96
|
14
|
0.145833333333333
|
0.0166666666666667
|
|
GO:BP
|
GO:0032102
|
negative regulation of response to external stimulus
|
0.000992510213766247
|
406
|
51
|
7
|
0.137254901960784
|
0.0172413793103448
|
|
GO:BP
|
GO:1901987
|
regulation of cell cycle phase transition
|
0.00101959671965628
|
493
|
88
|
10
|
0.113636363636364
|
0.0202839756592292
|
|
GO:BP
|
GO:0034248
|
regulation of cellular amide metabolic process
|
0.00103269259612536
|
535
|
28
|
6
|
0.214285714285714
|
0.011214953271028
|
|
GO:BP
|
GO:0071407
|
cellular response to organic cyclic compound
|
0.00103269259612536
|
587
|
120
|
13
|
0.108333333333333
|
0.0221465076660988
|
|
GO:BP
|
GO:0070647
|
protein modification by small protein conjugation or removal
|
0.00108463811488138
|
1184
|
96
|
17
|
0.177083333333333
|
0.0143581081081081
|
|
GO:BP
|
GO:0034620
|
cellular response to unfolded protein
|
0.00110034296394398
|
149
|
33
|
4
|
0.121212121212121
|
0.0268456375838926
|
|
GO:BP
|
GO:2000060
|
positive regulation of ubiquitin-dependent protein catabolic process
|
0.00114002439736108
|
107
|
46
|
4
|
0.0869565217391304
|
0.0373831775700935
|
|
GO:BP
|
GO:0010632
|
regulation of epithelial cell migration
|
0.00114523394202918
|
306
|
116
|
9
|
0.0775862068965517
|
0.0294117647058824
|
|
GO:BP
|
GO:0071214
|
cellular response to abiotic stimulus
|
0.00117733032214649
|
339
|
83
|
8
|
0.0963855421686747
|
0.023598820058997
|
|
GO:BP
|
GO:0001525
|
angiogenesis
|
0.00117733032214649
|
617
|
116
|
13
|
0.112068965517241
|
0.0210696920583468
|
|
GO:BP
|
GO:0104004
|
cellular response to environmental stimulus
|
0.00117733032214649
|
339
|
83
|
8
|
0.0963855421686747
|
0.023598820058997
|
|
GO:BP
|
GO:0007155
|
cell adhesion
|
0.00118881590713844
|
1492
|
57
|
14
|
0.245614035087719
|
0.00938337801608579
|
|
GO:BP
|
GO:0022610
|
biological adhesion
|
0.00124460862114709
|
1499
|
57
|
14
|
0.245614035087719
|
0.00933955970647098
|
|
GO:BP
|
GO:0007050
|
cell cycle arrest
|
0.00124460862114709
|
241
|
88
|
7
|
0.0795454545454545
|
0.029045643153527
|
|
GO:BP
|
GO:0006928
|
movement of cell or subcellular component
|
0.00124940472139548
|
2311
|
62
|
19
|
0.306451612903226
|
0.00822154911293812
|
|
GO:BP
|
GO:0010594
|
regulation of endothelial cell migration
|
0.00126722157362225
|
244
|
116
|
8
|
0.0689655172413793
|
0.0327868852459016
|
|
GO:BP
|
GO:0007585
|
respiratory gaseous exchange by respiratory system
|
0.00130651485063507
|
69
|
74
|
4
|
0.0540540540540541
|
0.0579710144927536
|
|
GO:BP
|
GO:1901800
|
positive regulation of proteasomal protein catabolic process
|
0.00132924358701081
|
112
|
46
|
4
|
0.0869565217391304
|
0.0357142857142857
|
|
GO:BP
|
GO:0006954
|
inflammatory response
|
0.0013662923151912
|
794
|
58
|
10
|
0.172413793103448
|
0.0125944584382872
|
|
GO:BP
|
GO:0009057
|
macromolecule catabolic process
|
0.00138375144064031
|
1468
|
33
|
10
|
0.303030303030303
|
0.00681198910081744
|
|
GO:BP
|
GO:0030218
|
erythrocyte differentiation
|
0.00138456091005221
|
116
|
45
|
4
|
0.0888888888888889
|
0.0344827586206897
|
|
GO:BP
|
GO:0007610
|
behavior
|
0.00138618469225887
|
586
|
10
|
4
|
0.4
|
0.0068259385665529
|
|
GO:BP
|
GO:0042493
|
response to drug
|
0.00140202968020834
|
396
|
6
|
3
|
0.5
|
0.00757575757575758
|
|
GO:BP
|
GO:0042770
|
signal transduction in response to DNA damage
|
0.00142820137442559
|
135
|
113
|
6
|
0.0530973451327434
|
0.0444444444444444
|
|
GO:BP
|
GO:0022414
|
reproductive process
|
0.0014509096357646
|
1493
|
94
|
19
|
0.202127659574468
|
0.0127260549229739
|
|
GO:BP
|
GO:0030509
|
BMP signaling pathway
|
0.0014784842098869
|
158
|
34
|
4
|
0.117647058823529
|
0.0253164556962025
|
|
GO:BP
|
GO:0000003
|
reproduction
|
0.0014784842098869
|
1496
|
94
|
19
|
0.202127659574468
|
0.0127005347593583
|
|
GO:BP
|
GO:0022407
|
regulation of cell-cell adhesion
|
0.00158055464067078
|
445
|
123
|
11
|
0.0894308943089431
|
0.0247191011235955
|
|
GO:BP
|
GO:1903409
|
reactive oxygen species biosynthetic process
|
0.00158158519303549
|
135
|
74
|
5
|
0.0675675675675676
|
0.037037037037037
|
|
GO:BP
|
GO:2000134
|
negative regulation of G1/S transition of mitotic cell cycle
|
0.00158158519303549
|
128
|
78
|
5
|
0.0641025641025641
|
0.0390625
|
|
GO:BP
|
GO:0007369
|
gastrulation
|
0.00161550926411786
|
185
|
120
|
7
|
0.0583333333333333
|
0.0378378378378378
|
|
GO:BP
|
GO:0044772
|
mitotic cell cycle phase transition
|
0.00162729934839222
|
609
|
122
|
13
|
0.10655737704918
|
0.0213464696223317
|
|
GO:BP
|
GO:0045620
|
negative regulation of lymphocyte differentiation
|
0.00162729934839222
|
57
|
96
|
4
|
0.0416666666666667
|
0.0701754385964912
|
|
GO:BP
|
GO:0009611
|
response to wounding
|
0.00162729934839222
|
674
|
57
|
9
|
0.157894736842105
|
0.013353115727003
|
|
GO:BP
|
GO:0035967
|
cellular response to topologically incorrect protein
|
0.00162850464532872
|
168
|
33
|
4
|
0.121212121212121
|
0.0238095238095238
|
|
GO:BP
|
GO:0035019
|
somatic stem cell population maintenance
|
0.00162850464532872
|
72
|
76
|
4
|
0.0526315789473684
|
0.0555555555555556
|
|
GO:BP
|
GO:0071398
|
cellular response to fatty acid
|
0.00164277815819647
|
43
|
51
|
3
|
0.0588235294117647
|
0.0697674418604651
|
|
GO:BP
|
GO:0007159
|
leukocyte cell-cell adhesion
|
0.00165231813713235
|
375
|
123
|
10
|
0.0813008130081301
|
0.0266666666666667
|
|
GO:BP
|
GO:0050673
|
epithelial cell proliferation
|
0.00165261124788616
|
460
|
120
|
11
|
0.0916666666666667
|
0.0239130434782609
|
|
GO:BP
|
GO:1902533
|
positive regulation of intracellular signal transduction
|
0.00165261124788616
|
1037
|
83
|
14
|
0.168674698795181
|
0.0135004821600771
|
|
GO:BP
|
GO:0044843
|
cell cycle G1/S phase transition
|
0.00167260127462565
|
309
|
122
|
9
|
0.0737704918032787
|
0.029126213592233
|
|
GO:BP
|
GO:0034101
|
erythrocyte homeostasis
|
0.00169963363880454
|
124
|
45
|
4
|
0.0888888888888889
|
0.032258064516129
|
|
GO:BP
|
GO:0001667
|
ameboidal-type cell migration
|
0.00180102257059221
|
494
|
78
|
9
|
0.115384615384615
|
0.0182186234817814
|
|
GO:BP
|
GO:0032872
|
regulation of stress-activated MAPK cascade
|
0.0018269689383918
|
190
|
85
|
6
|
0.0705882352941176
|
0.0315789473684211
|
|
GO:BP
|
GO:0045429
|
positive regulation of nitric oxide biosynthetic process
|
0.0018494261615076
|
45
|
51
|
3
|
0.0588235294117647
|
0.0666666666666667
|
|
GO:BP
|
GO:1902807
|
negative regulation of cell cycle G1/S phase transition
|
0.00189241793369005
|
134
|
78
|
5
|
0.0641025641025641
|
0.0373134328358209
|
|
GO:BP
|
GO:0060674
|
placenta blood vessel development
|
0.00189241793369005
|
31
|
75
|
3
|
0.04
|
0.0967741935483871
|
|
GO:BP
|
GO:0043367
|
CD4-positive, alpha-beta T cell differentiation
|
0.00189435958998081
|
85
|
123
|
5
|
0.040650406504065
|
0.0588235294117647
|
|
GO:BP
|
GO:0071772
|
response to BMP
|
0.00190236952373049
|
171
|
34
|
4
|
0.117647058823529
|
0.0233918128654971
|
|
GO:BP
|
GO:0071773
|
cellular response to BMP stimulus
|
0.00190236952373049
|
171
|
34
|
4
|
0.117647058823529
|
0.0233918128654971
|
|
GO:BP
|
GO:1904407
|
positive regulation of nitric oxide metabolic process
|
0.00194512295722503
|
46
|
51
|
3
|
0.0588235294117647
|
0.0652173913043478
|
|
GO:BP
|
GO:0070302
|
regulation of stress-activated protein kinase signaling cascade
|
0.00195029909308783
|
193
|
85
|
6
|
0.0705882352941176
|
0.0310880829015544
|
|
GO:BP
|
GO:0030334
|
regulation of cell migration
|
0.00205607169481318
|
999
|
120
|
17
|
0.141666666666667
|
0.017017017017017
|
|
GO:BP
|
GO:0007254
|
JNK cascade
|
0.00205607169481318
|
195
|
85
|
6
|
0.0705882352941176
|
0.0307692307692308
|
|
GO:BP
|
GO:0007611
|
learning or memory
|
0.00207307761515089
|
257
|
10
|
3
|
0.3
|
0.0116731517509728
|
|
GO:BP
|
GO:1900745
|
positive regulation of p38MAPK cascade
|
0.00208155469072564
|
31
|
78
|
3
|
0.0384615384615385
|
0.0967741935483871
|
|
GO:BP
|
GO:0045732
|
positive regulation of protein catabolic process
|
0.00213108732507788
|
237
|
46
|
5
|
0.108695652173913
|
0.0210970464135021
|
|
GO:BP
|
GO:0033002
|
muscle cell proliferation
|
0.00213108732507788
|
244
|
25
|
4
|
0.16
|
0.0163934426229508
|
|
GO:BP
|
GO:0050678
|
regulation of epithelial cell proliferation
|
0.00219129036006545
|
401
|
120
|
10
|
0.0833333333333333
|
0.0249376558603491
|
|
GO:BP
|
GO:1903052
|
positive regulation of proteolysis involved in cellular protein catabolic process
|
0.00219129036006545
|
131
|
46
|
4
|
0.0869565217391304
|
0.0305343511450382
|
|
GO:BP
|
GO:0090303
|
positive regulation of wound healing
|
0.00219129036006545
|
60
|
41
|
3
|
0.0731707317073171
|
0.05
|
|
GO:BP
|
GO:0032101
|
regulation of response to external stimulus
|
0.00223936947869485
|
1051
|
75
|
13
|
0.173333333333333
|
0.0123691722169363
|
|
GO:BP
|
GO:0022402
|
cell cycle process
|
0.00233374441663337
|
1439
|
24
|
8
|
0.333333333333333
|
0.00555941626129256
|
|
GO:BP
|
GO:0016071
|
mRNA metabolic process
|
0.00235587013960674
|
874
|
29
|
7
|
0.241379310344828
|
0.0080091533180778
|
|
GO:BP
|
GO:0042327
|
positive regulation of phosphorylation
|
0.00235587013960674
|
927
|
96
|
14
|
0.145833333333333
|
0.0151024811218986
|
|
GO:BP
|
GO:0007162
|
negative regulation of cell adhesion
|
0.00236152082180425
|
304
|
104
|
8
|
0.0769230769230769
|
0.0263157894736842
|
|
GO:BP
|
GO:0032434
|
regulation of proteasomal ubiquitin-dependent protein catabolic process
|
0.00236237907630943
|
134
|
46
|
4
|
0.0869565217391304
|
0.0298507462686567
|
|
GO:BP
|
GO:0033674
|
positive regulation of kinase activity
|
0.00237469056640161
|
608
|
96
|
11
|
0.114583333333333
|
0.0180921052631579
|
|
GO:BP
|
GO:0002753
|
cytoplasmic pattern recognition receptor signaling pathway
|
0.00240159720007575
|
73
|
35
|
3
|
0.0857142857142857
|
0.0410958904109589
|
|
GO:BP
|
GO:0030335
|
positive regulation of cell migration
|
0.00245279844541115
|
567
|
120
|
12
|
0.1
|
0.0211640211640212
|
|
GO:BP
|
GO:0051707
|
response to other organism
|
0.00248578154089843
|
1592
|
58
|
14
|
0.241379310344828
|
0.00879396984924623
|
|
GO:BP
|
GO:0043370
|
regulation of CD4-positive, alpha-beta T cell differentiation
|
0.00251126689748579
|
51
|
123
|
4
|
0.032520325203252
|
0.0784313725490196
|
|
GO:BP
|
GO:0043207
|
response to external biotic stimulus
|
0.00251126689748579
|
1594
|
58
|
14
|
0.241379310344828
|
0.00878293601003764
|
|
GO:BP
|
GO:0010883
|
regulation of lipid storage
|
0.00259988760002693
|
56
|
113
|
4
|
0.0353982300884956
|
0.0714285714285714
|
|
GO:BP
|
GO:0097305
|
response to alcohol
|
0.00259988760002693
|
255
|
45
|
5
|
0.111111111111111
|
0.0196078431372549
|
|
GO:BP
|
GO:0010608
|
posttranscriptional regulation of gene expression
|
0.00267452984532421
|
896
|
29
|
7
|
0.241379310344828
|
0.0078125
|
|
GO:BP
|
GO:0043281
|
regulation of cysteine-type endopeptidase activity involved in apoptotic process
|
0.00267452984532421
|
217
|
113
|
7
|
0.0619469026548673
|
0.032258064516129
|
|
GO:BP
|
GO:0034976
|
response to endoplasmic reticulum stress
|
0.00272091136128402
|
298
|
83
|
7
|
0.0843373493975904
|
0.023489932885906
|
|
GO:BP
|
GO:0044342
|
type B pancreatic cell proliferation
|
0.00284028114951782
|
23
|
120
|
3
|
0.025
|
0.130434782608696
|
|
GO:BP
|
GO:1903362
|
regulation of cellular protein catabolic process
|
0.00293125497427622
|
257
|
46
|
5
|
0.108695652173913
|
0.0194552529182879
|
|
GO:BP
|
GO:0009607
|
response to biotic stimulus
|
0.00300110426672315
|
1626
|
58
|
14
|
0.241379310344828
|
0.00861008610086101
|
|
GO:BP
|
GO:0050890
|
cognition
|
0.00300953781161492
|
298
|
10
|
3
|
0.3
|
0.0100671140939597
|
|
GO:BP
|
GO:0001824
|
blastocyst development
|
0.00301029698951114
|
104
|
27
|
3
|
0.111111111111111
|
0.0288461538461538
|
|
GO:BP
|
GO:0030856
|
regulation of epithelial cell differentiation
|
0.00303203302603969
|
164
|
72
|
5
|
0.0694444444444444
|
0.0304878048780488
|
|
GO:BP
|
GO:0044770
|
cell cycle phase transition
|
0.00304652397823306
|
658
|
122
|
13
|
0.10655737704918
|
0.0197568389057751
|
|
GO:BP
|
GO:0045600
|
positive regulation of fat cell differentiation
|
0.00308292582236565
|
67
|
42
|
3
|
0.0714285714285714
|
0.0447761194029851
|
|
GO:BP
|
GO:0097193
|
intrinsic apoptotic signaling pathway
|
0.00315510243506846
|
294
|
113
|
8
|
0.0707964601769911
|
0.0272108843537415
|
|
GO:BP
|
GO:0044321
|
response to leptin
|
0.00318035958968605
|
24
|
120
|
3
|
0.025
|
0.125
|
|
GO:BP
|
GO:0010564
|
regulation of cell cycle process
|
0.0032136354471808
|
809
|
24
|
6
|
0.25
|
0.00741656365883807
|
|
GO:BP
|
GO:0002262
|
myeloid cell homeostasis
|
0.00324007778603136
|
151
|
45
|
4
|
0.0888888888888889
|
0.0264900662251656
|
|
GO:BP
|
GO:0044248
|
cellular catabolic process
|
0.00337218673112498
|
2347
|
33
|
12
|
0.363636363636364
|
0.00511291009799744
|
|
GO:BP
|
GO:2000147
|
positive regulation of cell motility
|
0.00337394558887005
|
591
|
120
|
12
|
0.1
|
0.0203045685279188
|
|
GO:BP
|
GO:0030155
|
regulation of cell adhesion
|
0.00337394558887005
|
748
|
123
|
14
|
0.113821138211382
|
0.018716577540107
|
|
GO:BP
|
GO:0022603
|
regulation of anatomical structure morphogenesis
|
0.00339505925599095
|
1042
|
51
|
10
|
0.196078431372549
|
0.00959692898272553
|
|
GO:BP
|
GO:0051270
|
regulation of cellular component movement
|
0.00340590579340252
|
1141
|
121
|
18
|
0.148760330578512
|
0.0157756354075372
|
|
GO:BP
|
GO:0008625
|
extrinsic apoptotic signaling pathway via death domain receptors
|
0.00345010641179917
|
83
|
83
|
4
|
0.0481927710843374
|
0.0481927710843374
|
|
GO:BP
|
GO:0051345
|
positive regulation of hydrolase activity
|
0.00345010641179917
|
783
|
118
|
14
|
0.11864406779661
|
0.0178799489144317
|
|
GO:BP
|
GO:0007179
|
transforming growth factor beta receptor signaling pathway
|
0.00347144059845274
|
206
|
34
|
4
|
0.117647058823529
|
0.0194174757281553
|
|
GO:BP
|
GO:0061041
|
regulation of wound healing
|
0.00347321220871373
|
136
|
51
|
4
|
0.0784313725490196
|
0.0294117647058824
|
|
GO:BP
|
GO:0022408
|
negative regulation of cell-cell adhesion
|
0.00349345335050059
|
194
|
96
|
6
|
0.0625
|
0.0309278350515464
|
|
GO:BP
|
GO:0032446
|
protein modification by small protein conjugation
|
0.00354260521995927
|
973
|
96
|
14
|
0.145833333333333
|
0.0143884892086331
|
|
GO:BP
|
GO:0006970
|
response to osmotic stress
|
0.00357378887946803
|
84
|
83
|
4
|
0.0481927710843374
|
0.0476190476190476
|
|
GO:BP
|
GO:0001704
|
formation of primary germ layer
|
0.00357378887946803
|
123
|
100
|
5
|
0.05
|
0.040650406504065
|
|
GO:BP
|
GO:0051085
|
chaperone cofactor-dependent protein refolding
|
0.00357692310966947
|
31
|
97
|
3
|
0.0309278350515464
|
0.0967741935483871
|
|
GO:BP
|
GO:2000145
|
regulation of cell motility
|
0.00358679267635566
|
1061
|
42
|
9
|
0.214285714285714
|
0.00848256361922714
|
|
GO:BP
|
GO:0033077
|
T cell differentiation in thymus
|
0.00358679267635566
|
76
|
92
|
4
|
0.0434782608695652
|
0.0526315789473684
|
|
GO:BP
|
GO:0002040
|
sprouting angiogenesis
|
0.00358679267635566
|
204
|
92
|
6
|
0.0652173913043478
|
0.0294117647058824
|
|
GO:BP
|
GO:0002694
|
regulation of leukocyte activation
|
0.00360291229349697
|
610
|
57
|
8
|
0.140350877192982
|
0.0131147540983607
|
|
GO:BP
|
GO:0048661
|
positive regulation of smooth muscle cell proliferation
|
0.00361880536023668
|
103
|
120
|
5
|
0.0416666666666667
|
0.0485436893203883
|
|
GO:BP
|
GO:1903036
|
positive regulation of response to wounding
|
0.00367044710207911
|
74
|
41
|
3
|
0.0731707317073171
|
0.0405405405405405
|
|
GO:BP
|
GO:0046649
|
lymphocyte activation
|
0.00368910892454292
|
771
|
57
|
9
|
0.157894736842105
|
0.0116731517509728
|
|
GO:BP
|
GO:0045616
|
regulation of keratinocyte differentiation
|
0.00373084420911343
|
43
|
71
|
3
|
0.0422535211267606
|
0.0697674418604651
|
|
GO:BP
|
GO:0031396
|
regulation of protein ubiquitination
|
0.00375217206981402
|
211
|
90
|
6
|
0.0666666666666667
|
0.028436018957346
|
|
GO:BP
|
GO:0070059
|
intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress
|
0.00375729523850974
|
63
|
113
|
4
|
0.0353982300884956
|
0.0634920634920635
|
|
GO:BP
|
GO:0046634
|
regulation of alpha-beta T cell activation
|
0.00381553596873594
|
102
|
123
|
5
|
0.040650406504065
|
0.0490196078431373
|
|
GO:BP
|
GO:0046631
|
alpha-beta T cell activation
|
0.00382037926048976
|
155
|
123
|
6
|
0.0487804878048781
|
0.0387096774193548
|
|
GO:BP
|
GO:0051272
|
positive regulation of cellular component movement
|
0.0038603377515483
|
604
|
120
|
12
|
0.1
|
0.0198675496688742
|
|
GO:BP
|
GO:0040017
|
positive regulation of locomotion
|
0.00396498562221552
|
606
|
120
|
12
|
0.1
|
0.0198019801980198
|
|
GO:BP
|
GO:0030855
|
epithelial cell differentiation
|
0.00396498562221552
|
798
|
45
|
8
|
0.177777777777778
|
0.0100250626566416
|
|
GO:BP
|
GO:1900034
|
regulation of cellular response to heat
|
0.00401133328411506
|
79
|
40
|
3
|
0.075
|
0.0379746835443038
|
|
GO:BP
|
GO:0030183
|
B cell differentiation
|
0.00406146787780466
|
140
|
23
|
3
|
0.130434782608696
|
0.0214285714285714
|
|
GO:BP
|
GO:0051704
|
multi-organism process
|
0.00406146787780466
|
1135
|
94
|
15
|
0.159574468085106
|
0.013215859030837
|
|
GO:BP
|
GO:0035710
|
CD4-positive, alpha-beta T cell activation
|
0.00406146787780466
|
104
|
123
|
5
|
0.040650406504065
|
0.0480769230769231
|
|
GO:BP
|
GO:1903203
|
regulation of oxidative stress-induced neuron death
|
0.00417772639457892
|
27
|
120
|
3
|
0.025
|
0.111111111111111
|
|
GO:BP
|
GO:0045321
|
leukocyte activation
|
0.00427407625062674
|
1332
|
57
|
12
|
0.210526315789474
|
0.00900900900900901
|
|
GO:BP
|
GO:0043491
|
protein kinase B signaling
|
0.00431197070455609
|
286
|
46
|
5
|
0.108695652173913
|
0.0174825174825175
|
|
GO:BP
|
GO:2000116
|
regulation of cysteine-type endopeptidase activity
|
0.0043397292155537
|
240
|
113
|
7
|
0.0619469026548673
|
0.0291666666666667
|
|
GO:BP
|
GO:0030336
|
negative regulation of cell migration
|
0.0044087975292303
|
353
|
22
|
4
|
0.181818181818182
|
0.0113314447592068
|
|
GO:BP
|
GO:0001890
|
placenta development
|
0.00442335930203163
|
147
|
89
|
5
|
0.0561797752808989
|
0.0340136054421769
|
|
GO:BP
|
GO:2000058
|
regulation of ubiquitin-dependent protein catabolic process
|
0.00449859836370663
|
165
|
46
|
4
|
0.0869565217391304
|
0.0242424242424242
|
|
GO:BP
|
GO:0007167
|
enzyme linked receptor protein signaling pathway
|
0.00449936415640801
|
1103
|
34
|
8
|
0.235294117647059
|
0.00725294650951949
|
|
GO:BP
|
GO:0071470
|
cellular response to osmotic stress
|
0.00451655193136391
|
40
|
83
|
3
|
0.036144578313253
|
0.075
|
|
GO:BP
|
GO:0022604
|
regulation of cell morphogenesis
|
0.00469170309580595
|
309
|
116
|
8
|
0.0689655172413793
|
0.0258899676375405
|
|
GO:BP
|
GO:0040013
|
negative regulation of locomotion
|
0.00474822446180526
|
401
|
90
|
8
|
0.0888888888888889
|
0.0199501246882793
|
|
GO:BP
|
GO:0071158
|
positive regulation of cell cycle arrest
|
0.00487233663098095
|
88
|
88
|
4
|
0.0454545454545455
|
0.0454545454545455
|
|
GO:BP
|
GO:0043153
|
entrainment of circadian clock by photoperiod
|
0.00488230197454993
|
29
|
119
|
3
|
0.0252100840336134
|
0.103448275862069
|
|
GO:BP
|
GO:0048872
|
homeostasis of number of cells
|
0.00488513502643076
|
261
|
52
|
5
|
0.0961538461538462
|
0.0191570881226054
|
|
GO:BP
|
GO:0071479
|
cellular response to ionizing radiation
|
0.00494299606879032
|
69
|
113
|
4
|
0.0353982300884956
|
0.0579710144927536
|
|
GO:BP
|
GO:2000146
|
negative regulation of cell motility
|
0.00502699997307232
|
369
|
22
|
4
|
0.181818181818182
|
0.010840108401084
|
|
GO:BP
|
GO:0014065
|
phosphatidylinositol 3-kinase signaling
|
0.00506233488599009
|
161
|
22
|
3
|
0.136363636363636
|
0.0186335403726708
|
|
GO:BP
|
GO:0051084
|
'de novo' posttranslational protein folding
|
0.00506233488599009
|
36
|
97
|
3
|
0.0309278350515464
|
0.0833333333333333
|
|
GO:BP
|
GO:0006412
|
translation
|
0.00506233488599009
|
764
|
28
|
6
|
0.214285714285714
|
0.00785340314136126
|
|
GO:BP
|
GO:1903038
|
negative regulation of leukocyte cell-cell adhesion
|
0.00513134760401547
|
142
|
96
|
5
|
0.0520833333333333
|
0.0352112676056338
|
|
GO:BP
|
GO:0050865
|
regulation of cell activation
|
0.00519915868491045
|
655
|
57
|
8
|
0.140350877192982
|
0.0122137404580153
|
|
GO:BP
|
GO:0070542
|
response to fatty acid
|
0.00519915868491045
|
69
|
51
|
3
|
0.0588235294117647
|
0.0434782608695652
|
|
GO:BP
|
GO:0001503
|
ossification
|
0.00532323753242021
|
417
|
110
|
9
|
0.0818181818181818
|
0.0215827338129496
|
|
GO:BP
|
GO:0051271
|
negative regulation of cellular component movement
|
0.00535449808152134
|
377
|
22
|
4
|
0.181818181818182
|
0.0106100795755968
|
|
GO:BP
|
GO:0043523
|
regulation of neuron apoptotic process
|
0.00535449808152134
|
216
|
17
|
3
|
0.176470588235294
|
0.0138888888888889
|
|
GO:BP
|
GO:0070498
|
interleukin-1-mediated signaling pathway
|
0.00574970771241817
|
105
|
35
|
3
|
0.0857142857142857
|
0.0285714285714286
|
|
GO:BP
|
GO:0036475
|
neuron death in response to oxidative stress
|
0.00587203711435918
|
31
|
120
|
3
|
0.025
|
0.0967741935483871
|
|
GO:BP
|
GO:0043043
|
peptide biosynthetic process
|
0.00587203711435918
|
790
|
28
|
6
|
0.214285714285714
|
0.00759493670886076
|
|
GO:BP
|
GO:2000514
|
regulation of CD4-positive, alpha-beta T cell activation
|
0.00587203711435918
|
67
|
123
|
4
|
0.032520325203252
|
0.0597014925373134
|
|
GO:BP
|
GO:0061448
|
connective tissue development
|
0.00590143794484432
|
256
|
113
|
7
|
0.0619469026548673
|
0.02734375
|
|
GO:BP
|
GO:1902235
|
regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway
|
0.00590706519277153
|
33
|
113
|
3
|
0.0265486725663717
|
0.0909090909090909
|
|
GO:BP
|
GO:0043407
|
negative regulation of MAP kinase activity
|
0.00601124338259206
|
80
|
104
|
4
|
0.0384615384615385
|
0.05
|
|
GO:BP
|
GO:0051249
|
regulation of lymphocyte activation
|
0.00601124338259206
|
518
|
57
|
7
|
0.12280701754386
|
0.0135135135135135
|
|
GO:BP
|
GO:0046637
|
regulation of alpha-beta T cell differentiation
|
0.00608912493942904
|
68
|
123
|
4
|
0.032520325203252
|
0.0588235294117647
|
|
GO:BP
|
GO:2001238
|
positive regulation of extrinsic apoptotic signaling pathway
|
0.00618456921334506
|
48
|
79
|
3
|
0.0379746835443038
|
0.0625
|
|
GO:BP
|
GO:0008593
|
regulation of Notch signaling pathway
|
0.00633982196075287
|
110
|
35
|
3
|
0.0857142857142857
|
0.0272727272727273
|
|
GO:BP
|
GO:0061136
|
regulation of proteasomal protein catabolic process
|
0.00641321399319232
|
186
|
46
|
4
|
0.0869565217391304
|
0.021505376344086
|
|
GO:BP
|
GO:0016032
|
viral process
|
0.00644705836994477
|
942
|
42
|
8
|
0.19047619047619
|
0.00849256900212314
|
|
GO:BP
|
GO:0042752
|
regulation of circadian rhythm
|
0.00644705836994477
|
122
|
119
|
5
|
0.0420168067226891
|
0.040983606557377
|
|
GO:BP
|
GO:2001235
|
positive regulation of apoptotic signaling pathway
|
0.00653593474121326
|
129
|
113
|
5
|
0.0442477876106195
|
0.0387596899224806
|
|
GO:BP
|
GO:0009648
|
photoperiodism
|
0.00661363830631188
|
33
|
119
|
3
|
0.0252100840336134
|
0.0909090909090909
|
|
GO:BP
|
GO:0090092
|
regulation of transmembrane receptor protein serine/threonine kinase signaling pathway
|
0.00663040909278587
|
257
|
34
|
4
|
0.117647058823529
|
0.0155642023346304
|
|
GO:BP
|
GO:0043372
|
positive regulation of CD4-positive, alpha-beta T cell differentiation
|
0.00663040909278587
|
32
|
123
|
3
|
0.024390243902439
|
0.09375
|
|
GO:BP
|
GO:0009649
|
entrainment of circadian clock
|
0.00710806203928904
|
34
|
119
|
3
|
0.0252100840336134
|
0.0882352941176471
|
|
GO:BP
|
GO:0052547
|
regulation of peptidase activity
|
0.00718719347431181
|
467
|
84
|
8
|
0.0952380952380952
|
0.0171306209850107
|
|
GO:BP
|
GO:0043516
|
regulation of DNA damage response, signal transduction by p53 class mediator
|
0.00722184170655905
|
36
|
113
|
3
|
0.0265486725663717
|
0.0833333333333333
|
|
GO:BP
|
GO:0006952
|
defense response
|
0.00722699640166753
|
1818
|
58
|
14
|
0.241379310344828
|
0.0077007700770077
|
|
GO:BP
|
GO:0009416
|
response to light stimulus
|
0.00723252198375837
|
328
|
119
|
8
|
0.0672268907563025
|
0.024390243902439
|
|
GO:BP
|
GO:0072331
|
signal transduction by p53 class mediator
|
0.00723252198375837
|
268
|
113
|
7
|
0.0619469026548673
|
0.0261194029850746
|
|
GO:BP
|
GO:0032088
|
negative regulation of NF-kappaB transcription factor activity
|
0.00726096607194135
|
97
|
42
|
3
|
0.0714285714285714
|
0.0309278350515464
|
|
GO:BP
|
GO:0051402
|
neuron apoptotic process
|
0.00753565452767214
|
250
|
17
|
3
|
0.176470588235294
|
0.012
|
|
GO:BP
|
GO:0090207
|
regulation of triglyceride metabolic process
|
0.00754349620323095
|
41
|
101
|
3
|
0.0297029702970297
|
0.0731707317073171
|
|
GO:BP
|
GO:0043462
|
regulation of ATPase activity
|
0.0076992113142803
|
80
|
113
|
4
|
0.0353982300884956
|
0.05
|
|
GO:BP
|
GO:2001237
|
negative regulation of extrinsic apoptotic signaling pathway
|
0.00779502315617221
|
108
|
84
|
4
|
0.0476190476190476
|
0.037037037037037
|
|
GO:BP
|
GO:0030968
|
endoplasmic reticulum unfolded protein response
|
0.00784218819123739
|
128
|
33
|
3
|
0.0909090909090909
|
0.0234375
|
|
GO:BP
|
GO:0010634
|
positive regulation of epithelial cell migration
|
0.0081014809021271
|
181
|
51
|
4
|
0.0784313725490196
|
0.0220994475138122
|
|
GO:BP
|
GO:0048015
|
phosphatidylinositol-mediated signaling
|
0.00811237095173106
|
197
|
22
|
3
|
0.136363636363636
|
0.0152284263959391
|
|
GO:BP
|
GO:0002009
|
morphogenesis of an epithelium
|
0.008201321042851
|
560
|
89
|
9
|
0.101123595505618
|
0.0160714285714286
|
|
GO:BP
|
GO:0044703
|
multi-organism reproductive process
|
0.00826115619399839
|
1040
|
5
|
3
|
0.6
|
0.00288461538461538
|
|
GO:BP
|
GO:0007622
|
rhythmic behavior
|
0.00828771399494881
|
49
|
88
|
3
|
0.0340909090909091
|
0.0612244897959184
|
|
GO:BP
|
GO:0002683
|
negative regulation of immune system process
|
0.00832049479644581
|
420
|
96
|
8
|
0.0833333333333333
|
0.019047619047619
|
|
GO:BP
|
GO:0048017
|
inositol lipid-mediated signaling
|
0.00834284458759312
|
201
|
22
|
3
|
0.136363636363636
|
0.0149253731343284
|
|
GO:BP
|
GO:0008285
|
negative regulation of cell population proliferation
|
0.00843346579953917
|
790
|
41
|
7
|
0.170731707317073
|
0.00886075949367089
|
|
GO:BP
|
GO:1901222
|
regulation of NIK/NF-kappaB signaling
|
0.00845944239006123
|
113
|
83
|
4
|
0.0481927710843374
|
0.0353982300884956
|
|
GO:BP
|
GO:1905897
|
regulation of response to endoplasmic reticulum stress
|
0.00845944239006123
|
84
|
52
|
3
|
0.0576923076923077
|
0.0357142857142857
|
|
GO:BP
|
GO:0060612
|
adipose tissue development
|
0.00856938512221421
|
39
|
113
|
3
|
0.0265486725663717
|
0.0769230769230769
|
|
GO:BP
|
GO:0032874
|
positive regulation of stress-activated MAPK cascade
|
0.00860082473912505
|
121
|
78
|
4
|
0.0512820512820513
|
0.0330578512396694
|
|
GO:BP
|
GO:0007492
|
endoderm development
|
0.00860082473912505
|
80
|
55
|
3
|
0.0545454545454545
|
0.0375
|
|
GO:BP
|
GO:0044403
|
biological process involved in symbiotic interaction
|
0.00865978968474716
|
1001
|
42
|
8
|
0.19047619047619
|
0.00799200799200799
|
|
GO:BP
|
GO:1903201
|
regulation of oxidative stress-induced cell death
|
0.00869872909034213
|
79
|
120
|
4
|
0.0333333333333333
|
0.0506329113924051
|
|
GO:BP
|
GO:0046888
|
negative regulation of hormone secretion
|
0.00869872909034213
|
66
|
67
|
3
|
0.0447761194029851
|
0.0454545454545455
|
|
GO:BP
|
GO:0038034
|
signal transduction in absence of ligand
|
0.0089517255339969
|
71
|
63
|
3
|
0.0476190476190476
|
0.0422535211267606
|
|
GO:BP
|
GO:0097192
|
extrinsic apoptotic signaling pathway in absence of ligand
|
0.0089517255339969
|
71
|
63
|
3
|
0.0476190476190476
|
0.0422535211267606
|
|
GO:BP
|
GO:0070304
|
positive regulation of stress-activated protein kinase signaling cascade
|
0.00903747920820265
|
123
|
78
|
4
|
0.0512820512820513
|
0.032520325203252
|
|
GO:BP
|
GO:0038061
|
NIK/NF-kappaB signaling
|
0.00910380002207069
|
194
|
83
|
5
|
0.0602409638554217
|
0.0257731958762887
|
|
GO:BP
|
GO:0006977
|
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest
|
0.00918155821880863
|
58
|
78
|
3
|
0.0384615384615385
|
0.0517241379310345
|
|
GO:BP
|
GO:0042149
|
cellular response to glucose starvation
|
0.00933386140988918
|
48
|
95
|
3
|
0.0315789473684211
|
0.0625
|
|
GO:BP
|
GO:0001775
|
cell activation
|
0.00938363571905046
|
1495
|
57
|
12
|
0.210526315789474
|
0.00802675585284281
|
|
GO:BP
|
GO:0019915
|
lipid storage
|
0.00939029510491677
|
86
|
113
|
4
|
0.0353982300884956
|
0.0465116279069767
|
|
GO:BP
|
GO:0030323
|
respiratory tube development
|
0.00949315584201676
|
177
|
92
|
5
|
0.0543478260869565
|
0.0282485875706215
|
|
GO:BP
|
GO:0008284
|
positive regulation of cell population proliferation
|
0.00949315584201676
|
982
|
11
|
4
|
0.363636363636364
|
0.00407331975560081
|
|
GO:BP
|
GO:0050863
|
regulation of T cell activation
|
0.00950887595412451
|
337
|
96
|
7
|
0.0729166666666667
|
0.0207715133531157
|
|
GO:BP
|
GO:1902400
|
intracellular signal transduction involved in G1 DNA damage checkpoint
|
0.00950887595412451
|
59
|
78
|
3
|
0.0384615384615385
|
0.0508474576271186
|
|
GO:BP
|
GO:0072431
|
signal transduction involved in mitotic G1 DNA damage checkpoint
|
0.00950887595412451
|
59
|
78
|
3
|
0.0384615384615385
|
0.0508474576271186
|
|
GO:BP
|
GO:2000516
|
positive regulation of CD4-positive, alpha-beta T cell activation
|
0.00978975187079499
|
38
|
123
|
3
|
0.024390243902439
|
0.0789473684210526
|
|
GO:BP
|
GO:0051336
|
regulation of hydrolase activity
|
0.0098786301143495
|
1321
|
118
|
18
|
0.152542372881356
|
0.0136260408781226
|
|
GO:BP
|
GO:0019827
|
stem cell population maintenance
|
0.0098786301143495
|
152
|
31
|
3
|
0.0967741935483871
|
0.0197368421052632
|
|
GO:BP
|
GO:0001816
|
cytokine production
|
0.00991223337152364
|
844
|
126
|
14
|
0.111111111111111
|
0.0165876777251185
|
|
GO:BP
|
GO:0034605
|
cellular response to heat
|
0.00996373969011801
|
118
|
40
|
3
|
0.075
|
0.0254237288135593
|
|
GO:BP
|
GO:0009056
|
catabolic process
|
0.0100282868586579
|
2744
|
33
|
12
|
0.363636363636364
|
0.0043731778425656
|
|
GO:BP
|
GO:0098727
|
maintenance of cell number
|
0.0101222173878485
|
154
|
31
|
3
|
0.0967741935483871
|
0.0194805194805195
|
|
GO:BP
|
GO:1902402
|
signal transduction involved in mitotic DNA damage checkpoint
|
0.0102539440414634
|
61
|
78
|
3
|
0.0384615384615385
|
0.0491803278688525
|
|
GO:BP
|
GO:1902403
|
signal transduction involved in mitotic DNA integrity checkpoint
|
0.0102539440414634
|
61
|
78
|
3
|
0.0384615384615385
|
0.0491803278688525
|
|
GO:BP
|
GO:0007249
|
I-kappaB kinase/NF-kappaB signaling
|
0.0103091289942997
|
291
|
113
|
7
|
0.0619469026548673
|
0.0240549828178694
|
|
GO:BP
|
GO:1990830
|
cellular response to leukemia inhibitory factor
|
0.010313599855949
|
89
|
113
|
4
|
0.0353982300884956
|
0.0449438202247191
|
|
GO:BP
|
GO:1903050
|
regulation of proteolysis involved in cellular protein catabolic process
|
0.0104034295839748
|
221
|
46
|
4
|
0.0869565217391304
|
0.0180995475113122
|
|
GO:BP
|
GO:0071156
|
regulation of cell cycle arrest
|
0.0105066837535548
|
115
|
88
|
4
|
0.0454545454545455
|
0.0347826086956522
|
|
GO:BP
|
GO:1990823
|
response to leukemia inhibitory factor
|
0.0106495857926651
|
90
|
113
|
4
|
0.0353982300884956
|
0.0444444444444444
|
|
GO:BP
|
GO:0031347
|
regulation of defense response
|
0.0106857886475706
|
704
|
75
|
9
|
0.12
|
0.0127840909090909
|
|
GO:BP
|
GO:1902041
|
regulation of extrinsic apoptotic signaling pathway via death domain receptors
|
0.0109411523880922
|
59
|
83
|
3
|
0.036144578313253
|
0.0508474576271186
|
|
GO:BP
|
GO:0045685
|
regulation of glial cell differentiation
|
0.0109411523880922
|
72
|
68
|
3
|
0.0441176470588235
|
0.0416666666666667
|
|
GO:BP
|
GO:0032868
|
response to insulin
|
0.0109496159501766
|
295
|
113
|
7
|
0.0619469026548673
|
0.023728813559322
|
|
GO:BP
|
GO:0072413
|
signal transduction involved in mitotic cell cycle checkpoint
|
0.0110249554711911
|
63
|
78
|
3
|
0.0384615384615385
|
0.0476190476190476
|
|
GO:BP
|
GO:0043604
|
amide biosynthetic process
|
0.0112581900462199
|
932
|
28
|
6
|
0.214285714285714
|
0.00643776824034335
|
|
GO:BP
|
GO:0033574
|
response to testosterone
|
0.0112696769001225
|
44
|
113
|
3
|
0.0265486725663717
|
0.0681818181818182
|
|
GO:BP
|
GO:0046425
|
regulation of receptor signaling pathway via JAK-STAT
|
0.0112878710471832
|
108
|
96
|
4
|
0.0416666666666667
|
0.037037037037037
|
|
GO:BP
|
GO:0007219
|
Notch signaling pathway
|
0.0113559536287365
|
193
|
89
|
5
|
0.0561797752808989
|
0.0259067357512953
|
|
GO:BP
|
GO:0010498
|
proteasomal protein catabolic process
|
0.0117227777184398
|
491
|
52
|
6
|
0.115384615384615
|
0.0122199592668024
|
|
GO:BP
|
GO:0070098
|
chemokine-mediated signaling pathway
|
0.0117335462116547
|
87
|
58
|
3
|
0.0517241379310345
|
0.0344827586206897
|
|
GO:BP
|
GO:0031571
|
mitotic G1 DNA damage checkpoint
|
0.0122875039405775
|
66
|
78
|
3
|
0.0384615384615385
|
0.0454545454545455
|
|
GO:BP
|
GO:0006518
|
peptide metabolic process
|
0.0124333906545749
|
955
|
28
|
6
|
0.214285714285714
|
0.00628272251308901
|
|
GO:BP
|
GO:0002221
|
pattern recognition receptor signaling pathway
|
0.0125248208177588
|
213
|
83
|
5
|
0.0602409638554217
|
0.0234741784037559
|
|
GO:BP
|
GO:0050679
|
positive regulation of epithelial cell proliferation
|
0.0125248208177588
|
213
|
120
|
6
|
0.05
|
0.028169014084507
|
|
GO:BP
|
GO:0035264
|
multicellular organism growth
|
0.0126103202272825
|
140
|
77
|
4
|
0.051948051948052
|
0.0285714285714286
|
|
GO:BP
|
GO:0006935
|
chemotaxis
|
0.0126484261307015
|
653
|
68
|
8
|
0.117647058823529
|
0.0122511485451761
|
|
GO:BP
|
GO:0046777
|
protein autophosphorylation
|
0.012672913599007
|
240
|
107
|
6
|
0.0560747663551402
|
0.025
|
|
GO:BP
|
GO:0044819
|
mitotic G1/S transition checkpoint
|
0.012672913599007
|
67
|
78
|
3
|
0.0384615384615385
|
0.0447761194029851
|
|
GO:BP
|
GO:0044783
|
G1 DNA damage checkpoint
|
0.012672913599007
|
67
|
78
|
3
|
0.0384615384615385
|
0.0447761194029851
|
|
GO:BP
|
GO:0042330
|
taxis
|
0.01290822515618
|
656
|
68
|
8
|
0.117647058823529
|
0.0121951219512195
|
|
GO:BP
|
GO:0050730
|
regulation of peptidyl-tyrosine phosphorylation
|
0.01290822515618
|
269
|
96
|
6
|
0.0625
|
0.0223048327137546
|
|
GO:BP
|
GO:1900407
|
regulation of cellular response to oxidative stress
|
0.0131118766164982
|
91
|
120
|
4
|
0.0333333333333333
|
0.043956043956044
|
|
GO:BP
|
GO:1901655
|
cellular response to ketone
|
0.0132665245348934
|
97
|
113
|
4
|
0.0353982300884956
|
0.0412371134020619
|
|
GO:BP
|
GO:0048729
|
tissue morphogenesis
|
0.0133937828223647
|
667
|
114
|
11
|
0.0964912280701754
|
0.0164917541229385
|
|
GO:BP
|
GO:0002695
|
negative regulation of leukocyte activation
|
0.0135117105694048
|
188
|
96
|
5
|
0.0520833333333333
|
0.0265957446808511
|
|
GO:BP
|
GO:0050727
|
regulation of inflammatory response
|
0.0135421747749539
|
381
|
48
|
5
|
0.104166666666667
|
0.0131233595800525
|
|
GO:BP
|
GO:0009913
|
epidermal cell differentiation
|
0.01358024325131
|
365
|
72
|
6
|
0.0833333333333333
|
0.0164383561643836
|
|
GO:BP
|
GO:0071902
|
positive regulation of protein serine/threonine kinase activity
|
0.0136119874074428
|
316
|
83
|
6
|
0.072289156626506
|
0.0189873417721519
|
|
GO:BP
|
GO:0050728
|
negative regulation of inflammatory response
|
0.0138168854342465
|
171
|
32
|
3
|
0.09375
|
0.0175438596491228
|
|
GO:BP
|
GO:0042176
|
regulation of protein catabolic process
|
0.0138273622493126
|
401
|
46
|
5
|
0.108695652173913
|
0.0124688279301746
|
|
GO:BP
|
GO:0043406
|
positive regulation of MAP kinase activity
|
0.013864624915737
|
234
|
78
|
5
|
0.0641025641025641
|
0.0213675213675214
|
|
GO:BP
|
GO:0065008
|
regulation of biological quality
|
0.0139102601678468
|
4131
|
52
|
21
|
0.403846153846154
|
0.00508351488743646
|
|
GO:BP
|
GO:0045582
|
positive regulation of T cell differentiation
|
0.0139354493597285
|
91
|
123
|
4
|
0.032520325203252
|
0.043956043956044
|
|
GO:BP
|
GO:0045787
|
positive regulation of cell cycle
|
0.01401459354199
|
403
|
88
|
7
|
0.0795454545454545
|
0.0173697270471464
|
|
GO:BP
|
GO:0002684
|
positive regulation of immune system process
|
0.01401459354199
|
1065
|
53
|
9
|
0.169811320754717
|
0.00845070422535211
|
|
GO:BP
|
GO:0098742
|
cell-cell adhesion via plasma-membrane adhesion molecules
|
0.0140354786458039
|
277
|
41
|
4
|
0.0975609756097561
|
0.0144404332129964
|
|
GO:BP
|
GO:0002042
|
cell migration involved in sprouting angiogenesis
|
0.0141010446611216
|
104
|
53
|
3
|
0.0566037735849057
|
0.0288461538461538
|
|
GO:BP
|
GO:0016925
|
protein sumoylation
|
0.0141068528426564
|
81
|
68
|
3
|
0.0441176470588235
|
0.037037037037037
|
|
GO:BP
|
GO:0040007
|
growth
|
0.0141628435620002
|
967
|
116
|
14
|
0.120689655172414
|
0.0144777662874871
|
|
GO:BP
|
GO:0032781
|
positive regulation of ATPase activity
|
0.0141650284412047
|
57
|
97
|
3
|
0.0309278350515464
|
0.0526315789473684
|
|
GO:BP
|
GO:0046328
|
regulation of JNK cascade
|
0.0142310572556283
|
133
|
85
|
4
|
0.0470588235294118
|
0.0300751879699248
|
|
GO:BP
|
GO:0006974
|
cellular response to DNA damage stimulus
|
0.0142999992638954
|
888
|
52
|
8
|
0.153846153846154
|
0.00900900900900901
|
|
GO:BP
|
GO:1904892
|
regulation of receptor signaling pathway via STAT
|
0.0143265983172917
|
118
|
96
|
4
|
0.0416666666666667
|
0.0338983050847458
|
|
GO:BP
|
GO:0006820
|
anion transport
|
0.0143519022708596
|
2889
|
77
|
22
|
0.285714285714286
|
0.00761509172724126
|
|
GO:BP
|
GO:0043551
|
regulation of phosphatidylinositol 3-kinase activity
|
0.0143519022708596
|
58
|
96
|
3
|
0.03125
|
0.0517241379310345
|
|
GO:BP
|
GO:0071897
|
DNA biosynthetic process
|
0.0144662066087142
|
195
|
29
|
3
|
0.103448275862069
|
0.0153846153846154
|
|
GO:BP
|
GO:0050732
|
negative regulation of peptidyl-tyrosine phosphorylation
|
0.0148025916417497
|
59
|
96
|
3
|
0.03125
|
0.0508474576271186
|
|
GO:BP
|
GO:0090068
|
positive regulation of cell cycle process
|
0.0153667409304743
|
309
|
88
|
6
|
0.0681818181818182
|
0.0194174757281553
|
|
GO:BP
|
GO:0055088
|
lipid homeostasis
|
0.0153787288974197
|
167
|
113
|
5
|
0.0442477876106195
|
0.029940119760479
|
|
GO:BP
|
GO:1905954
|
positive regulation of lipid localization
|
0.0153787288974197
|
113
|
51
|
3
|
0.0588235294117647
|
0.0265486725663717
|
|
GO:BP
|
GO:0060759
|
regulation of response to cytokine stimulus
|
0.0153787288974197
|
197
|
96
|
5
|
0.0520833333333333
|
0.0253807106598985
|
|
GO:BP
|
GO:0001817
|
regulation of cytokine production
|
0.0156139100706126
|
806
|
126
|
13
|
0.103174603174603
|
0.0161290322580645
|
|
GO:BP
|
GO:0003158
|
endothelium development
|
0.0158905243361248
|
143
|
41
|
3
|
0.0731707317073171
|
0.020979020979021
|
|
GO:BP
|
GO:0072422
|
signal transduction involved in DNA damage checkpoint
|
0.0159454997024746
|
75
|
78
|
3
|
0.0384615384615385
|
0.04
|
|
GO:BP
|
GO:0007422
|
peripheral nervous system development
|
0.0159454997024746
|
79
|
74
|
3
|
0.0405405405405405
|
0.0379746835443038
|
|
GO:BP
|
GO:0072401
|
signal transduction involved in DNA integrity checkpoint
|
0.0159454997024746
|
75
|
78
|
3
|
0.0384615384615385
|
0.04
|
|
GO:BP
|
GO:0050868
|
negative regulation of T cell activation
|
0.0159513439541992
|
123
|
96
|
4
|
0.0416666666666667
|
0.032520325203252
|
|
GO:BP
|
GO:1904707
|
positive regulation of vascular associated smooth muscle cell proliferation
|
0.0160242448644267
|
49
|
120
|
3
|
0.025
|
0.0612244897959184
|
|
GO:BP
|
GO:0097435
|
supramolecular fiber organization
|
0.0162490280699418
|
734
|
51
|
7
|
0.137254901960784
|
0.00953678474114441
|
|
GO:BP
|
GO:0040008
|
regulation of growth
|
0.0163551312835303
|
681
|
116
|
11
|
0.0948275862068965
|
0.0161527165932452
|
|
GO:BP
|
GO:0018108
|
peptidyl-tyrosine phosphorylation
|
0.0166116708535331
|
385
|
96
|
7
|
0.0729166666666667
|
0.0181818181818182
|
|
GO:BP
|
GO:0030593
|
neutrophil chemotaxis
|
0.0166296441278611
|
103
|
58
|
3
|
0.0517241379310345
|
0.029126213592233
|
|
GO:BP
|
GO:0021915
|
neural tube development
|
0.0166296441278611
|
158
|
38
|
3
|
0.0789473684210526
|
0.0189873417721519
|
|
GO:BP
|
GO:1902882
|
regulation of response to oxidative stress
|
0.0166296441278611
|
100
|
120
|
4
|
0.0333333333333333
|
0.04
|
|
GO:BP
|
GO:0036473
|
cell death in response to oxidative stress
|
0.0166296441278611
|
100
|
120
|
4
|
0.0333333333333333
|
0.04
|
|
GO:BP
|
GO:0051179
|
localization
|
0.0168944374119098
|
6901
|
68
|
37
|
0.544117647058823
|
0.00536154180553543
|
|
GO:BP
|
GO:1900024
|
regulation of substrate adhesion-dependent cell spreading
|
0.0169649509326636
|
58
|
104
|
3
|
0.0288461538461538
|
0.0517241379310345
|
|
GO:BP
|
GO:0018212
|
peptidyl-tyrosine modification
|
0.0171416015165636
|
388
|
96
|
7
|
0.0729166666666667
|
0.0180412371134021
|
|
GO:BP
|
GO:0007568
|
aging
|
0.0171623544385461
|
329
|
113
|
7
|
0.0619469026548673
|
0.0212765957446809
|
|
GO:BP
|
GO:0071705
|
nitrogen compound transport
|
0.0171798984371623
|
2322
|
68
|
17
|
0.25
|
0.00732127476313523
|
|
GO:BP
|
GO:0072395
|
signal transduction involved in cell cycle checkpoint
|
0.0172245880724526
|
78
|
78
|
3
|
0.0384615384615385
|
0.0384615384615385
|
|
GO:BP
|
GO:0043161
|
proteasome-mediated ubiquitin-dependent protein catabolic process
|
0.0174855223496048
|
434
|
46
|
5
|
0.108695652173913
|
0.0115207373271889
|
|
GO:BP
|
GO:0009791
|
post-embryonic development
|
0.0175500558500463
|
84
|
73
|
3
|
0.0410958904109589
|
0.0357142857142857
|
|
GO:BP
|
GO:0046638
|
positive regulation of alpha-beta T cell differentiation
|
0.0175576665358955
|
50
|
123
|
3
|
0.024390243902439
|
0.06
|
|
GO:BP
|
GO:0048568
|
embryonic organ development
|
0.0177990054731906
|
432
|
110
|
8
|
0.0727272727272727
|
0.0185185185185185
|
|
GO:BP
|
GO:0071260
|
cellular response to mechanical stimulus
|
0.0181290369431752
|
80
|
78
|
3
|
0.0384615384615385
|
0.0375
|
|
GO:BP
|
GO:0006508
|
proteolysis
|
0.0181625204107701
|
1836
|
52
|
12
|
0.230769230769231
|
0.0065359477124183
|
|
GO:BP
|
GO:0071621
|
granulocyte chemotaxis
|
0.0183335076672157
|
126
|
99
|
4
|
0.0404040404040404
|
0.0317460317460317
|
|
GO:BP
|
GO:0000902
|
cell morphogenesis
|
0.0183484449634284
|
1037
|
68
|
10
|
0.147058823529412
|
0.00964320154291225
|
|
GO:BP
|
GO:0010676
|
positive regulation of cellular carbohydrate metabolic process
|
0.0184234472025768
|
56
|
113
|
3
|
0.0265486725663717
|
0.0535714285714286
|
|
GO:BP
|
GO:0051098
|
regulation of binding
|
0.0185185404707034
|
365
|
35
|
4
|
0.114285714285714
|
0.010958904109589
|
|
GO:BP
|
GO:0043502
|
regulation of muscle adaptation
|
0.0186607175572086
|
105
|
120
|
4
|
0.0333333333333333
|
0.0380952380952381
|
|
GO:BP
|
GO:0050866
|
negative regulation of cell activation
|
0.0188150922483379
|
211
|
96
|
5
|
0.0520833333333333
|
0.023696682464455
|
|
GO:BP
|
GO:0060070
|
canonical Wnt signaling pathway
|
0.0189379238970403
|
345
|
84
|
6
|
0.0714285714285714
|
0.0173913043478261
|
|
GO:BP
|
GO:0001649
|
osteoblast differentiation
|
0.0189681194537318
|
232
|
28
|
3
|
0.107142857142857
|
0.0129310344827586
|
|
GO:BP
|
GO:1901796
|
regulation of signal transduction by p53 class mediator
|
0.019155073484852
|
180
|
113
|
5
|
0.0442477876106195
|
0.0277777777777778
|
|
GO:BP
|
GO:0097529
|
myeloid leukocyte migration
|
0.0192743666793366
|
221
|
58
|
4
|
0.0689655172413793
|
0.0180995475113122
|
|
GO:BP
|
GO:0006641
|
triglyceride metabolic process
|
0.0193096809308634
|
113
|
113
|
4
|
0.0353982300884956
|
0.0353982300884956
|
|
GO:BP
|
GO:0001938
|
positive regulation of endothelial cell proliferation
|
0.0193096809308634
|
113
|
113
|
4
|
0.0353982300884956
|
0.0353982300884956
|
|
GO:BP
|
GO:0045621
|
positive regulation of lymphocyte differentiation
|
0.0193541499536305
|
104
|
123
|
4
|
0.032520325203252
|
0.0384615384615385
|
|
GO:BP
|
GO:1901575
|
organic substance catabolic process
|
0.0195854802998674
|
2267
|
33
|
10
|
0.303030303030303
|
0.00441111601235112
|
|
GO:BP
|
GO:1901566
|
organonitrogen compound biosynthetic process
|
0.0196385240381879
|
1853
|
28
|
8
|
0.285714285714286
|
0.00431732325957906
|
|
GO:BP
|
GO:0016567
|
protein ubiquitination
|
0.0199902680372479
|
892
|
119
|
13
|
0.109243697478992
|
0.0145739910313901
|
|
GO:BP
|
GO:0070372
|
regulation of ERK1 and ERK2 cascade
|
0.0205815365644291
|
309
|
22
|
3
|
0.136363636363636
|
0.00970873786407767
|
|
GO:BP
|
GO:0030595
|
leukocyte chemotaxis
|
0.0207005085803196
|
227
|
58
|
4
|
0.0689655172413793
|
0.0176211453744493
|
|
GO:BP
|
GO:0045926
|
negative regulation of growth
|
0.0207005085803196
|
255
|
116
|
6
|
0.0517241379310345
|
0.0235294117647059
|
|
GO:BP
|
GO:0042592
|
homeostatic process
|
0.0207720153485699
|
1981
|
101
|
20
|
0.198019801980198
|
0.0100959111559818
|
|
GO:BP
|
GO:0046330
|
positive regulation of JNK cascade
|
0.0208683894604872
|
86
|
78
|
3
|
0.0384615384615385
|
0.0348837209302326
|
|
GO:BP
|
GO:1903039
|
positive regulation of leukocyte cell-cell adhesion
|
0.0208683894604872
|
241
|
123
|
6
|
0.0487804878048781
|
0.024896265560166
|
|
GO:BP
|
GO:0043550
|
regulation of lipid kinase activity
|
0.0209383815453605
|
70
|
96
|
3
|
0.03125
|
0.0428571428571429
|
|
GO:BP
|
GO:0007498
|
mesoderm development
|
0.0210570049954311
|
132
|
100
|
4
|
0.04
|
0.0303030303030303
|
|
GO:BP
|
GO:0043534
|
blood vessel endothelial cell migration
|
0.0212200006149121
|
186
|
113
|
5
|
0.0442477876106195
|
0.0268817204301075
|
|
GO:BP
|
GO:0050729
|
positive regulation of inflammatory response
|
0.0214429759206222
|
142
|
48
|
3
|
0.0625
|
0.0211267605633803
|
|
GO:BP
|
GO:0072594
|
establishment of protein localization to organelle
|
0.0215079804169742
|
584
|
52
|
6
|
0.115384615384615
|
0.0102739726027397
|
|
GO:BP
|
GO:0002224
|
toll-like receptor signaling pathway
|
0.0217235368941525
|
161
|
83
|
4
|
0.0481927710843374
|
0.0248447204968944
|
|
GO:BP
|
GO:0003002
|
regionalization
|
0.0217235368941525
|
338
|
89
|
6
|
0.0674157303370786
|
0.0177514792899408
|
|
GO:BP
|
GO:0010951
|
negative regulation of endopeptidase activity
|
0.0217988295524998
|
253
|
84
|
5
|
0.0595238095238095
|
0.0197628458498024
|
|
GO:BP
|
GO:0014013
|
regulation of gliogenesis
|
0.0219366794603537
|
101
|
68
|
3
|
0.0441176470588235
|
0.0297029702970297
|
|
GO:BP
|
GO:1903202
|
negative regulation of oxidative stress-induced cell death
|
0.0224649137902722
|
58
|
120
|
3
|
0.025
|
0.0517241379310345
|
|
GO:BP
|
GO:0051128
|
regulation of cellular component organization
|
0.0224649137902722
|
2412
|
52
|
14
|
0.269230769230769
|
0.00580431177446103
|
|
GO:BP
|
GO:0019216
|
regulation of lipid metabolic process
|
0.0226059136329601
|
419
|
120
|
8
|
0.0666666666666667
|
0.0190930787589499
|
|
GO:BP
|
GO:2001244
|
positive regulation of intrinsic apoptotic signaling pathway
|
0.0226287185456342
|
62
|
113
|
3
|
0.0265486725663717
|
0.0483870967741935
|
|
GO:BP
|
GO:0010595
|
positive regulation of endothelial cell migration
|
0.0227708585692306
|
138
|
51
|
3
|
0.0588235294117647
|
0.0217391304347826
|
|
GO:BP
|
GO:0070830
|
bicellular tight junction assembly
|
0.0234984404229602
|
80
|
89
|
3
|
0.0337078651685393
|
0.0375
|
|
GO:BP
|
GO:0006606
|
protein import into nucleus
|
0.0234984404229602
|
152
|
47
|
3
|
0.0638297872340425
|
0.0197368421052632
|
|
GO:BP
|
GO:0006457
|
protein folding
|
0.0236637474590015
|
225
|
97
|
5
|
0.0515463917525773
|
0.0222222222222222
|
|
GO:BP
|
GO:1990266
|
neutrophil migration
|
0.0239554241725261
|
124
|
58
|
3
|
0.0517241379310345
|
0.0241935483870968
|
|
GO:BP
|
GO:0051055
|
negative regulation of lipid biosynthetic process
|
0.0241996809649435
|
64
|
113
|
3
|
0.0265486725663717
|
0.046875
|
|
GO:BP
|
GO:0031330
|
negative regulation of cellular catabolic process
|
0.0241996809649435
|
262
|
28
|
3
|
0.107142857142857
|
0.0114503816793893
|
|
GO:BP
|
GO:0032651
|
regulation of interleukin-1 beta production
|
0.0242035298972834
|
102
|
71
|
3
|
0.0422535211267606
|
0.0294117647058824
|
|
GO:BP
|
GO:0010508
|
positive regulation of autophagy
|
0.0243953658235443
|
124
|
113
|
4
|
0.0353982300884956
|
0.032258064516129
|
|
GO:BP
|
GO:0007163
|
establishment or maintenance of cell polarity
|
0.0244376131564759
|
220
|
64
|
4
|
0.0625
|
0.0181818181818182
|
|
GO:BP
|
GO:0120192
|
tight junction assembly
|
0.0245867129672085
|
82
|
89
|
3
|
0.0337078651685393
|
0.0365853658536585
|
|
GO:BP
|
GO:0030308
|
negative regulation of cell growth
|
0.0251494251142448
|
192
|
116
|
5
|
0.0431034482758621
|
0.0260416666666667
|
|
GO:BP
|
GO:0001707
|
mesoderm formation
|
0.0253005530350056
|
74
|
100
|
3
|
0.03
|
0.0405405405405405
|
|
GO:BP
|
GO:0010466
|
negative regulation of peptidase activity
|
0.0254809484595369
|
267
|
84
|
5
|
0.0595238095238095
|
0.0187265917602996
|
|
GO:BP
|
GO:0042113
|
B cell activation
|
0.0256366368375557
|
330
|
23
|
3
|
0.130434782608696
|
0.00909090909090909
|
|
GO:BP
|
GO:0044257
|
cellular protein catabolic process
|
0.0259482050514505
|
813
|
52
|
7
|
0.134615384615385
|
0.00861008610086101
|
|
GO:BP
|
GO:0045765
|
regulation of angiogenesis
|
0.0259743197666321
|
358
|
116
|
7
|
0.0603448275862069
|
0.0195530726256983
|
|
GO:BP
|
GO:0032147
|
activation of protein kinase activity
|
0.0261026180905364
|
331
|
96
|
6
|
0.0625
|
0.0181268882175227
|
|
GO:BP
|
GO:0008544
|
epidermis development
|
0.0262123853343888
|
475
|
31
|
4
|
0.129032258064516
|
0.00842105263157895
|
|
GO:BP
|
GO:0032922
|
circadian regulation of gene expression
|
0.0262948338694527
|
67
|
113
|
3
|
0.0265486725663717
|
0.0447761194029851
|
|
GO:BP
|
GO:0043410
|
positive regulation of MAPK cascade
|
0.026328917687425
|
539
|
78
|
7
|
0.0897435897435897
|
0.012987012987013
|
|
GO:BP
|
GO:1905952
|
regulation of lipid localization
|
0.026380770710088
|
201
|
113
|
5
|
0.0442477876106195
|
0.0248756218905473
|
|
GO:BP
|
GO:0032611
|
interleukin-1 beta production
|
0.0263987972321797
|
107
|
71
|
3
|
0.0422535211267606
|
0.0280373831775701
|
|
GO:BP
|
GO:0014823
|
response to activity
|
0.0263987972321797
|
76
|
100
|
3
|
0.03
|
0.0394736842105263
|
|
GO:BP
|
GO:0048332
|
mesoderm morphogenesis
|
0.0263987972321797
|
76
|
100
|
3
|
0.03
|
0.0394736842105263
|
|
GO:BP
|
GO:0018193
|
peptidyl-amino acid modification
|
0.0264101035445663
|
1303
|
96
|
14
|
0.145833333333333
|
0.0107444359171144
|
|
GO:BP
|
GO:0048469
|
cell maturation
|
0.0265291895681443
|
170
|
45
|
3
|
0.0666666666666667
|
0.0176470588235294
|
|
GO:BP
|
GO:0042093
|
T-helper cell differentiation
|
0.026776552834553
|
66
|
116
|
3
|
0.0258620689655172
|
0.0454545454545455
|
|
GO:BP
|
GO:0019058
|
viral life cycle
|
0.0269836623719859
|
353
|
42
|
4
|
0.0952380952380952
|
0.0113314447592068
|
|
GO:BP
|
GO:0019218
|
regulation of steroid metabolic process
|
0.0271228453656433
|
130
|
113
|
4
|
0.0353982300884956
|
0.0307692307692308
|
|
GO:BP
|
GO:0000079
|
regulation of cyclin-dependent protein serine/threonine kinase activity
|
0.0271545708633221
|
99
|
78
|
3
|
0.0384615384615385
|
0.0303030303030303
|
|
GO:BP
|
GO:1901342
|
regulation of vasculature development
|
0.0272074683676371
|
364
|
116
|
7
|
0.0603448275862069
|
0.0192307692307692
|
|
GO:BP
|
GO:0120193
|
tight junction organization
|
0.0272967975276142
|
87
|
89
|
3
|
0.0337078651685393
|
0.0344827586206897
|
|
GO:BP
|
GO:0031348
|
negative regulation of defense response
|
0.0277260756759035
|
246
|
32
|
3
|
0.09375
|
0.0121951219512195
|
|
GO:BP
|
GO:1900076
|
regulation of cellular response to insulin stimulus
|
0.0277297653605623
|
69
|
113
|
3
|
0.0265486725663717
|
0.0434782608695652
|
|
GO:BP
|
GO:0045766
|
positive regulation of angiogenesis
|
0.0278226693447361
|
187
|
42
|
3
|
0.0714285714285714
|
0.0160427807486631
|
|
GO:BP
|
GO:1904018
|
positive regulation of vasculature development
|
0.0278226693447361
|
187
|
42
|
3
|
0.0714285714285714
|
0.0160427807486631
|
|
GO:BP
|
GO:0010952
|
positive regulation of peptidase activity
|
0.0278318922133745
|
205
|
113
|
5
|
0.0442477876106195
|
0.024390243902439
|
|
GO:BP
|
GO:0043297
|
apical junction assembly
|
0.0279715121780935
|
88
|
89
|
3
|
0.0337078651685393
|
0.0340909090909091
|
|
GO:BP
|
GO:0097530
|
granulocyte migration
|
0.0283021901717891
|
151
|
99
|
4
|
0.0404040404040404
|
0.0264900662251656
|
|
GO:BP
|
GO:0002294
|
CD4-positive, alpha-beta T cell differentiation involved in immune response
|
0.0283021901717891
|
68
|
116
|
3
|
0.0258620689655172
|
0.0441176470588235
|
|
GO:BP
|
GO:0030900
|
forebrain development
|
0.0283137736545547
|
385
|
61
|
5
|
0.0819672131147541
|
0.012987012987013
|
|
GO:BP
|
GO:0051250
|
negative regulation of lymphocyte activation
|
0.0283797162547279
|
156
|
96
|
4
|
0.0416666666666667
|
0.0256410256410256
|
|
GO:BP
|
GO:0062012
|
regulation of small molecule metabolic process
|
0.0290904379608021
|
447
|
120
|
8
|
0.0666666666666667
|
0.0178970917225951
|
|
GO:BP
|
GO:0002293
|
alpha-beta T cell differentiation involved in immune response
|
0.0291838903330946
|
69
|
116
|
3
|
0.0258620689655172
|
0.0434782608695652
|
|
GO:BP
|
GO:0002287
|
alpha-beta T cell activation involved in immune response
|
0.0291838903330946
|
69
|
116
|
3
|
0.0258620689655172
|
0.0434782608695652
|
|
GO:BP
|
GO:0046626
|
regulation of insulin receptor signaling pathway
|
0.0292994512562333
|
71
|
113
|
3
|
0.0265486725663717
|
0.0422535211267606
|
|
GO:BP
|
GO:0045785
|
positive regulation of cell adhesion
|
0.0292994512562333
|
437
|
123
|
8
|
0.0650406504065041
|
0.0183066361556064
|
|
GO:BP
|
GO:1904029
|
regulation of cyclin-dependent protein kinase activity
|
0.0293468544765073
|
103
|
78
|
3
|
0.0384615384615385
|
0.029126213592233
|
|
GO:BP
|
GO:0051170
|
import into nucleus
|
0.0295194499446829
|
172
|
47
|
3
|
0.0638297872340425
|
0.0174418604651163
|
|
GO:BP
|
GO:0051346
|
negative regulation of hydrolase activity
|
0.0296360294986148
|
470
|
33
|
4
|
0.121212121212121
|
0.00851063829787234
|
|
GO:BP
|
GO:0043500
|
muscle adaptation
|
0.0297014365670916
|
127
|
120
|
4
|
0.0333333333333333
|
0.031496062992126
|
|
GO:BP
|
GO:0071702
|
organic substance transport
|
0.0298717309206256
|
2766
|
77
|
20
|
0.25974025974026
|
0.00723065798987708
|
|
GO:BP
|
GO:0043603
|
cellular amide metabolic process
|
0.0304823899482109
|
1241
|
28
|
6
|
0.214285714285714
|
0.0048348106365834
|
|
GO:BP
|
GO:0045165
|
cell fate commitment
|
0.03059844689023
|
267
|
31
|
3
|
0.0967741935483871
|
0.0112359550561798
|
|
GO:BP
|
GO:0046635
|
positive regulation of alpha-beta T cell activation
|
0.0308781728803201
|
67
|
123
|
3
|
0.024390243902439
|
0.0447761194029851
|
|
GO:BP
|
GO:0021953
|
central nervous system neuron differentiation
|
0.0308781728803201
|
188
|
44
|
3
|
0.0681818181818182
|
0.0159574468085106
|
|
GO:BP
|
GO:0070727
|
cellular macromolecule localization
|
0.0309120825658419
|
2024
|
52
|
12
|
0.230769230769231
|
0.00592885375494071
|
|
GO:BP
|
GO:0010596
|
negative regulation of endothelial cell migration
|
0.031004942040937
|
106
|
78
|
3
|
0.0384615384615385
|
0.0283018867924528
|
|
GO:BP
|
GO:0044773
|
mitotic DNA damage checkpoint
|
0.031004942040937
|
106
|
78
|
3
|
0.0384615384615385
|
0.0283018867924528
|
|
GO:BP
|
GO:0035051
|
cardiocyte differentiation
|
0.0317399358644007
|
157
|
100
|
4
|
0.04
|
0.0254777070063694
|
|
GO:BP
|
GO:0042692
|
muscle cell differentiation
|
0.0317399358644007
|
390
|
113
|
7
|
0.0619469026548673
|
0.0179487179487179
|
|
GO:BP
|
GO:0046890
|
regulation of lipid biosynthetic process
|
0.0317399358644007
|
215
|
113
|
5
|
0.0442477876106195
|
0.0232558139534884
|
|
GO:BP
|
GO:0042509
|
regulation of tyrosine phosphorylation of STAT protein
|
0.0317399358644007
|
87
|
96
|
3
|
0.03125
|
0.0344827586206897
|
|
GO:BP
|
GO:0016055
|
Wnt signaling pathway
|
0.0320849044152565
|
529
|
84
|
7
|
0.0833333333333333
|
0.0132325141776938
|
|
GO:BP
|
GO:0032652
|
regulation of interleukin-1 production
|
0.0321674979941831
|
119
|
71
|
3
|
0.0422535211267606
|
0.0252100840336134
|
|
GO:BP
|
GO:0060333
|
interferon-gamma-mediated signaling pathway
|
0.0321807029841641
|
88
|
96
|
3
|
0.03125
|
0.0340909090909091
|
|
GO:BP
|
GO:0016579
|
protein deubiquitination
|
0.0323744879187007
|
292
|
84
|
5
|
0.0595238095238095
|
0.0171232876712329
|
|
GO:BP
|
GO:0198738
|
cell-cell signaling by wnt
|
0.0325272220396655
|
531
|
84
|
7
|
0.0833333333333333
|
0.0131826741996234
|
|
GO:BP
|
GO:0030324
|
lung development
|
0.0327127723330797
|
173
|
92
|
4
|
0.0434782608695652
|
0.023121387283237
|
|
GO:BP
|
GO:0007399
|
nervous system development
|
0.0328417994736673
|
2449
|
44
|
12
|
0.272727272727273
|
0.00489995916700694
|
|
GO:BP
|
GO:0006639
|
acylglycerol metabolic process
|
0.0328575420794902
|
141
|
113
|
4
|
0.0353982300884956
|
0.0283687943262411
|
|
GO:BP
|
GO:0007260
|
tyrosine phosphorylation of STAT protein
|
0.0328662200619372
|
89
|
96
|
3
|
0.03125
|
0.0337078651685393
|
|
GO:BP
|
GO:0032879
|
regulation of localization
|
0.032992990171323
|
2872
|
95
|
24
|
0.252631578947368
|
0.00835654596100279
|
|
GO:BP
|
GO:0006638
|
neutral lipid metabolic process
|
0.0334157110957273
|
142
|
113
|
4
|
0.0353982300884956
|
0.028169014084507
|
|
GO:BP
|
GO:0007267
|
cell-cell signaling
|
0.0335973435272052
|
1722
|
84
|
15
|
0.178571428571429
|
0.00871080139372822
|
|
GO:BP
|
GO:0061097
|
regulation of protein tyrosine kinase activity
|
0.0335973435272052
|
95
|
91
|
3
|
0.032967032967033
|
0.0315789473684211
|
|
GO:BP
|
GO:0002292
|
T cell differentiation involved in immune response
|
0.0339668661851279
|
75
|
116
|
3
|
0.0258620689655172
|
0.04
|
|
GO:BP
|
GO:0043588
|
skin development
|
0.0339751522276706
|
424
|
21
|
3
|
0.142857142857143
|
0.00707547169811321
|
|
GO:BP
|
GO:0051896
|
regulation of protein kinase B signaling
|
0.0340605759603905
|
259
|
34
|
3
|
0.0882352941176471
|
0.0115830115830116
|
|
GO:BP
|
GO:0044774
|
mitotic DNA integrity checkpoint
|
0.0341474825677349
|
112
|
78
|
3
|
0.0384615384615385
|
0.0267857142857143
|
|
GO:BP
|
GO:0033036
|
macromolecule localization
|
0.0341513468664696
|
3218
|
68
|
20
|
0.294117647058824
|
0.00621504039776259
|
|
GO:BP
|
GO:0006886
|
intracellular protein transport
|
0.0343685353475818
|
1186
|
68
|
10
|
0.147058823529412
|
0.00843170320404722
|
|
GO:BP
|
GO:0009895
|
negative regulation of catabolic process
|
0.0352516129863622
|
321
|
28
|
3
|
0.107142857142857
|
0.00934579439252336
|
|
GO:BP
|
GO:0022409
|
positive regulation of cell-cell adhesion
|
0.0352516129863622
|
284
|
123
|
6
|
0.0487804878048781
|
0.0211267605633803
|
|
GO:BP
|
GO:0010611
|
regulation of cardiac muscle hypertrophy
|
0.035362385993616
|
74
|
120
|
3
|
0.025
|
0.0405405405405405
|
|
GO:BP
|
GO:0003018
|
vascular process in circulatory system
|
0.0355731873279676
|
256
|
99
|
5
|
0.0505050505050505
|
0.01953125
|
|
GO:BP
|
GO:0032612
|
interleukin-1 production
|
0.0357540826773386
|
126
|
71
|
3
|
0.0422535211267606
|
0.0238095238095238
|
|
GO:BP
|
GO:0062207
|
regulation of pattern recognition receptor signaling pathway
|
0.0358546509159723
|
108
|
83
|
3
|
0.036144578313253
|
0.0277777777777778
|
|
GO:BP
|
GO:0015833
|
peptide transport
|
0.0360031456984469
|
1971
|
68
|
14
|
0.205882352941176
|
0.00710299340436327
|
|
GO:BP
|
GO:0006259
|
DNA metabolic process
|
0.036047364262671
|
965
|
10
|
3
|
0.3
|
0.00310880829015544
|
|
GO:BP
|
GO:0060562
|
epithelial tube morphogenesis
|
0.0360722322612261
|
329
|
51
|
4
|
0.0784313725490196
|
0.0121580547112462
|
|
GO:BP
|
GO:0045913
|
positive regulation of carbohydrate metabolic process
|
0.0361267063188851
|
80
|
113
|
3
|
0.0265486725663717
|
0.0375
|
|
GO:BP
|
GO:1901215
|
negative regulation of neuron death
|
0.0361544664207009
|
213
|
120
|
5
|
0.0416666666666667
|
0.0234741784037559
|
|
GO:BP
|
GO:0007259
|
receptor signaling pathway via JAK-STAT
|
0.0361785387975299
|
174
|
96
|
4
|
0.0416666666666667
|
0.0229885057471264
|
|
GO:BP
|
GO:0071346
|
cellular response to interferon-gamma
|
0.0364012541652762
|
175
|
96
|
4
|
0.0416666666666667
|
0.0228571428571429
|
|
GO:BP
|
GO:0010675
|
regulation of cellular carbohydrate metabolic process
|
0.0366337902965618
|
149
|
113
|
4
|
0.0353982300884956
|
0.0268456375838926
|
|
GO:BP
|
GO:2001020
|
regulation of response to DNA damage stimulus
|
0.0366337902965618
|
228
|
113
|
5
|
0.0442477876106195
|
0.0219298245614035
|
|
GO:BP
|
GO:1903793
|
positive regulation of anion transport
|
0.0371816355409762
|
504
|
52
|
5
|
0.0961538461538462
|
0.00992063492063492
|
|
GO:BP
|
GO:0014743
|
regulation of muscle hypertrophy
|
0.037219845425076
|
77
|
120
|
3
|
0.025
|
0.038961038961039
|
|
GO:BP
|
GO:0001818
|
negative regulation of cytokine production
|
0.0372605385532076
|
381
|
45
|
4
|
0.0888888888888889
|
0.010498687664042
|
|
GO:BP
|
GO:0007626
|
locomotory behavior
|
0.0373693113009322
|
189
|
90
|
4
|
0.0444444444444444
|
0.0211640211640212
|
|
GO:BP
|
GO:0070646
|
protein modification by small protein removal
|
0.0373693113009322
|
310
|
84
|
5
|
0.0595238095238095
|
0.0161290322580645
|
|
GO:BP
|
GO:0007507
|
heart development
|
0.0373899119960151
|
578
|
100
|
8
|
0.08
|
0.013840830449827
|
|
GO:BP
|
GO:0014032
|
neural crest cell development
|
0.0375284285648515
|
80
|
116
|
3
|
0.0258620689655172
|
0.0375
|
|
GO:BP
|
GO:0060284
|
regulation of cell development
|
0.038945269289657
|
502
|
116
|
8
|
0.0689655172413793
|
0.0159362549800797
|
|
GO:BP
|
GO:0002819
|
regulation of adaptive immune response
|
0.0394311107399697
|
167
|
57
|
3
|
0.0526315789473684
|
0.0179640718562874
|
|
GO:BP
|
GO:0042886
|
amide transport
|
0.0395030592447439
|
2007
|
68
|
14
|
0.205882352941176
|
0.00697558545092177
|
|
GO:BP
|
GO:0062013
|
positive regulation of small molecule metabolic process
|
0.0397014240888986
|
145
|
120
|
4
|
0.0333333333333333
|
0.0275862068965517
|
|
GO:BP
|
GO:0032869
|
cellular response to insulin stimulus
|
0.0399431244753589
|
235
|
113
|
5
|
0.0442477876106195
|
0.0212765957446809
|
|
GO:BP
|
GO:0052548
|
regulation of endopeptidase activity
|
0.0399431244753589
|
437
|
84
|
6
|
0.0714285714285714
|
0.0137299771167048
|
|
GO:BP
|
GO:0010633
|
negative regulation of epithelial cell migration
|
0.0402101361992404
|
123
|
78
|
3
|
0.0384615384615385
|
0.024390243902439
|
|
GO:BP
|
GO:0017038
|
protein import
|
0.0402101361992404
|
205
|
47
|
3
|
0.0638297872340425
|
0.0146341463414634
|
|
GO:BP
|
GO:0090316
|
positive regulation of intracellular protein transport
|
0.040604050167053
|
186
|
52
|
3
|
0.0576923076923077
|
0.0161290322580645
|
|
GO:BP
|
GO:0090287
|
regulation of cellular response to growth factor stimulus
|
0.0406993616729291
|
315
|
31
|
3
|
0.0967741935483871
|
0.00952380952380952
|
|
GO:BP
|
GO:0051129
|
negative regulation of cellular component organization
|
0.0407534530374022
|
759
|
36
|
5
|
0.138888888888889
|
0.00658761528326746
|
|
GO:BP
|
GO:0051493
|
regulation of cytoskeleton organization
|
0.0408225348594802
|
531
|
51
|
5
|
0.0980392156862745
|
0.00941619585687382
|
|
GO:BP
|
GO:0060021
|
roof of mouth development
|
0.0408225348594802
|
88
|
110
|
3
|
0.0272727272727273
|
0.0340909090909091
|
|
GO:BP
|
GO:0097696
|
receptor signaling pathway via STAT
|
0.0410202025194938
|
184
|
96
|
4
|
0.0416666666666667
|
0.0217391304347826
|
|
GO:BP
|
GO:0001959
|
regulation of cytokine-mediated signaling pathway
|
0.0410202025194938
|
184
|
96
|
4
|
0.0416666666666667
|
0.0217391304347826
|
|
GO:BP
|
GO:0014031
|
mesenchymal cell development
|
0.0412811652790977
|
84
|
116
|
3
|
0.0258620689655172
|
0.0357142857142857
|
|
GO:BP
|
GO:0048864
|
stem cell development
|
0.0412811652790977
|
84
|
116
|
3
|
0.0258620689655172
|
0.0357142857142857
|
|
GO:BP
|
GO:0050870
|
positive regulation of T cell activation
|
0.0414873104019012
|
219
|
123
|
5
|
0.040650406504065
|
0.0228310502283105
|
|
GO:BP
|
GO:0032370
|
positive regulation of lipid transport
|
0.041869150574721
|
87
|
113
|
3
|
0.0265486725663717
|
0.0344827586206897
|
|
GO:BP
|
GO:0050731
|
positive regulation of peptidyl-tyrosine phosphorylation
|
0.0429146019013934
|
198
|
91
|
4
|
0.043956043956044
|
0.0202020202020202
|
|
GO:BP
|
GO:0031214
|
biomineral tissue development
|
0.0430947180533501
|
164
|
110
|
4
|
0.0363636363636364
|
0.024390243902439
|
|
GO:BP
|
GO:0071695
|
anatomical structure maturation
|
0.0436085416550753
|
224
|
45
|
3
|
0.0666666666666667
|
0.0133928571428571
|
|
GO:BP
|
GO:0010720
|
positive regulation of cell development
|
0.0436150436335898
|
302
|
91
|
5
|
0.0549450549450549
|
0.0165562913907285
|
|
GO:BP
|
GO:0006094
|
gluconeogenesis
|
0.0437712584590001
|
89
|
113
|
3
|
0.0265486725663717
|
0.0337078651685393
|
|
GO:BP
|
GO:0090257
|
regulation of muscle system process
|
0.0439782117198132
|
257
|
71
|
4
|
0.0563380281690141
|
0.0155642023346304
|
|
GO:BP
|
GO:0010769
|
regulation of cell morphogenesis involved in differentiation
|
0.0439935131402301
|
97
|
104
|
3
|
0.0288461538461538
|
0.0309278350515464
|
|
GO:BP
|
GO:0042116
|
macrophage activation
|
0.0441750004776851
|
110
|
92
|
3
|
0.0326086956521739
|
0.0272727272727273
|
|
GO:BP
|
GO:0110148
|
biomineralization
|
0.0442147204130146
|
166
|
110
|
4
|
0.0363636363636364
|
0.0240963855421687
|
|
GO:BP
|
GO:0060541
|
respiratory system development
|
0.0442891042196649
|
199
|
92
|
4
|
0.0434782608695652
|
0.0201005025125628
|
|
GO:BP
|
GO:1903531
|
negative regulation of secretion by cell
|
0.0443866811486844
|
152
|
67
|
3
|
0.0447761194029851
|
0.0197368421052632
|
|
GO:BP
|
GO:0032956
|
regulation of actin cytoskeleton organization
|
0.0455526583776555
|
366
|
51
|
4
|
0.0784313725490196
|
0.0109289617486339
|
|
GO:BP
|
GO:0001558
|
regulation of cell growth
|
0.0460639670088833
|
426
|
116
|
7
|
0.0603448275862069
|
0.0164319248826291
|
|
GO:BP
|
GO:0019319
|
hexose biosynthetic process
|
0.046232502237673
|
92
|
113
|
3
|
0.0265486725663717
|
0.0326086956521739
|
|
GO:BP
|
GO:0051603
|
proteolysis involved in cellular protein catabolic process
|
0.0462894561862452
|
750
|
52
|
6
|
0.115384615384615
|
0.008
|
|
GO:BP
|
GO:0016525
|
negative regulation of angiogenesis
|
0.0463475688196991
|
161
|
116
|
4
|
0.0344827586206897
|
0.0248447204968944
|
|
GO:BP
|
GO:0014033
|
neural crest cell differentiation
|
0.0465295834878448
|
90
|
116
|
3
|
0.0258620689655172
|
0.0333333333333333
|
|
GO:BP
|
GO:0034341
|
response to interferon-gamma
|
0.046942158918713
|
196
|
96
|
4
|
0.0416666666666667
|
0.0204081632653061
|
|
GO:BP
|
GO:0030163
|
protein catabolic process
|
0.0470488752868687
|
974
|
52
|
7
|
0.134615384615385
|
0.00718685831622177
|
|
GO:BP
|
GO:1904705
|
regulation of vascular associated smooth muscle cell proliferation
|
0.0470488752868687
|
88
|
120
|
3
|
0.025
|
0.0340909090909091
|
|
GO:BP
|
GO:1990874
|
vascular associated smooth muscle cell proliferation
|
0.0470488752868687
|
88
|
120
|
3
|
0.025
|
0.0340909090909091
|
|
GO:BP
|
GO:0043122
|
regulation of I-kappaB kinase/NF-kappaB signaling
|
0.0470488752868687
|
251
|
113
|
5
|
0.0442477876106195
|
0.0199203187250996
|
|
GO:BP
|
GO:2000181
|
negative regulation of blood vessel morphogenesis
|
0.0472045592299442
|
163
|
116
|
4
|
0.0344827586206897
|
0.0245398773006135
|
|
GO:BP
|
GO:0010660
|
regulation of muscle cell apoptotic process
|
0.0475154756725491
|
94
|
113
|
3
|
0.0265486725663717
|
0.0319148936170213
|
|
GO:BP
|
GO:1901343
|
negative regulation of vasculature development
|
0.0478778356575754
|
164
|
116
|
4
|
0.0344827586206897
|
0.024390243902439
|
|
GO:BP
|
GO:0007389
|
pattern specification process
|
0.0487149846089276
|
442
|
89
|
6
|
0.0674157303370786
|
0.0135746606334842
|
|
GO:BP
|
GO:0042060
|
wound healing
|
0.0489947949458426
|
549
|
72
|
6
|
0.0833333333333333
|
0.0109289617486339
|
|
GO:CC
|
GO:0005634
|
nucleus
|
3.57218502332817e-11
|
7592
|
101
|
77
|
0.762376237623762
|
0.0101422550052687
|
|
GO:CC
|
GO:0035976
|
transcription factor AP-1 complex
|
9.82905517296626e-11
|
4
|
19
|
4
|
0.210526315789474
|
1
|
|
GO:CC
|
GO:0005654
|
nucleoplasm
|
1.79769805632811e-07
|
4102
|
101
|
49
|
0.485148514851485
|
0.0119453924914676
|
|
GO:CC
|
GO:0031981
|
nuclear lumen
|
6.72491489948046e-07
|
4444
|
101
|
50
|
0.495049504950495
|
0.0112511251125113
|
|
GO:CC
|
GO:0043231
|
intracellular membrane-bounded organelle
|
3.38998980585446e-06
|
11296
|
98
|
83
|
0.846938775510204
|
0.00734773371104816
|
|
GO:CC
|
GO:0000785
|
chromatin
|
7.31196785208873e-06
|
1526
|
8
|
7
|
0.875
|
0.00458715596330275
|
|
GO:CC
|
GO:0005694
|
chromosome
|
5.18955172328159e-05
|
2071
|
8
|
7
|
0.875
|
0.00338000965717045
|
|
GO:CC
|
GO:0070013
|
intracellular organelle lumen
|
7.36395467278683e-05
|
5402
|
101
|
51
|
0.504950495049505
|
0.00944094779711218
|
|
GO:CC
|
GO:0043229
|
intracellular organelle
|
7.36395467278683e-05
|
12491
|
98
|
85
|
0.86734693877551
|
0.00680489952765992
|
|
GO:CC
|
GO:0031974
|
membrane-enclosed lumen
|
0.000154579207859609
|
5565
|
101
|
51
|
0.504950495049505
|
0.0091644204851752
|
|
GO:CC
|
GO:0043233
|
organelle lumen
|
0.000154579207859609
|
5565
|
101
|
51
|
0.504950495049505
|
0.0091644204851752
|
|
GO:CC
|
GO:0005667
|
transcription regulator complex
|
0.000168297010591375
|
432
|
22
|
6
|
0.272727272727273
|
0.0138888888888889
|
|
GO:CC
|
GO:0005622
|
intracellular anatomical structure
|
0.00020746670186087
|
14790
|
101
|
95
|
0.940594059405941
|
0.00642325895875592
|
|
GO:CC
|
GO:0090575
|
RNA polymerase II transcription regulator complex
|
0.000337685129103295
|
183
|
6
|
3
|
0.5
|
0.0163934426229508
|
|
GO:CC
|
GO:0043227
|
membrane-bounded organelle
|
0.000362879121257329
|
12858
|
53
|
49
|
0.924528301886792
|
0.00381085705397418
|
|
GO:CC
|
GO:0043228
|
non-membrane-bounded organelle
|
0.000447292797115515
|
5346
|
101
|
48
|
0.475247524752475
|
0.00897867564534231
|
|
GO:CC
|
GO:0043232
|
intracellular non-membrane-bounded organelle
|
0.000447292797115515
|
5338
|
101
|
48
|
0.475247524752475
|
0.00899213188460097
|
|
GO:CC
|
GO:0000791
|
euchromatin
|
0.00154981550078418
|
41
|
113
|
4
|
0.0353982300884956
|
0.0975609756097561
|
|
GO:CC
|
GO:0043226
|
organelle
|
0.00186973304126122
|
14008
|
53
|
50
|
0.943396226415094
|
0.00356938892061679
|
|
GO:CC
|
GO:0005730
|
nucleolus
|
0.00893514194098788
|
955
|
92
|
13
|
0.141304347826087
|
0.0136125654450262
|
|
GO:CC
|
GO:0000932
|
P-body
|
0.0121973847274935
|
91
|
42
|
3
|
0.0714285714285714
|
0.032967032967033
|
|
GO:CC
|
GO:0036464
|
cytoplasmic ribonucleoprotein granule
|
0.0142046290320779
|
237
|
96
|
6
|
0.0625
|
0.0253164556962025
|
|
GO:CC
|
GO:0005737
|
cytoplasm
|
0.0157906937572465
|
11911
|
97
|
75
|
0.77319587628866
|
0.00629670052892284
|
|
GO:CC
|
GO:0016607
|
nuclear speck
|
0.0157906937572465
|
412
|
78
|
7
|
0.0897435897435897
|
0.0169902912621359
|
|
GO:CC
|
GO:0035770
|
ribonucleoprotein granule
|
0.0157906937572465
|
248
|
96
|
6
|
0.0625
|
0.0241935483870968
|
|
GO:CC
|
GO:0043657
|
host cell
|
0.0164701050638967
|
198
|
84
|
5
|
0.0595238095238095
|
0.0252525252525253
|
|
GO:CC
|
GO:0018995
|
host cellular component
|
0.0164701050638967
|
198
|
84
|
5
|
0.0595238095238095
|
0.0252525252525253
|
|
GO:CC
|
GO:0033648
|
host intracellular membrane-bounded organelle
|
0.018942847838474
|
143
|
124
|
5
|
0.0403225806451613
|
0.034965034965035
|
|
GO:CC
|
GO:0033647
|
host intracellular organelle
|
0.018942847838474
|
143
|
124
|
5
|
0.0403225806451613
|
0.034965034965035
|
|
GO:CC
|
GO:0005741
|
mitochondrial outer membrane
|
0.018942847838474
|
202
|
52
|
4
|
0.0769230769230769
|
0.0198019801980198
|
|
GO:CC
|
GO:0030666
|
endocytic vesicle membrane
|
0.018942847838474
|
159
|
66
|
4
|
0.0606060606060606
|
0.0251572327044025
|
|
GO:CC
|
GO:0042025
|
host cell nucleus
|
0.018942847838474
|
143
|
124
|
5
|
0.0403225806451613
|
0.034965034965035
|
|
GO:CC
|
GO:0043656
|
host intracellular region
|
0.0196320015654829
|
146
|
124
|
5
|
0.0403225806451613
|
0.0342465753424658
|
|
GO:CC
|
GO:0033646
|
host intracellular part
|
0.0196320015654829
|
146
|
124
|
5
|
0.0403225806451613
|
0.0342465753424658
|
|
GO:CC
|
GO:0033643
|
host cell part
|
0.0197002999123357
|
147
|
124
|
5
|
0.0403225806451613
|
0.0340136054421769
|
|
GO:CC
|
GO:0005829
|
cytosol
|
0.0224576974661744
|
5303
|
18
|
11
|
0.611111111111111
|
0.00207429756741467
|
|
GO:CC
|
GO:0031968
|
organelle outer membrane
|
0.0224582415461049
|
227
|
52
|
4
|
0.0769230769230769
|
0.0176211453744493
|
|
GO:CC
|
GO:0019867
|
outer membrane
|
0.02263358931308
|
229
|
52
|
4
|
0.0769230769230769
|
0.0174672489082969
|
|
GO:CC
|
GO:0140513
|
nuclear protein-containing complex
|
0.0289655060412848
|
1249
|
6
|
3
|
0.5
|
0.00240192153722978
|
|
GO:CC
|
GO:0032991
|
protein-containing complex
|
0.0379321977496695
|
5548
|
44
|
21
|
0.477272727272727
|
0.00378514780100937
|
|
GO:CC
|
GO:0005923
|
bicellular tight junction
|
0.0431181076782924
|
123
|
65
|
3
|
0.0461538461538462
|
0.024390243902439
|
|
GO:CC
|
GO:0001650
|
fibrillar center
|
0.0467501965769024
|
141
|
113
|
4
|
0.0353982300884956
|
0.0283687943262411
|
|
GO:CC
|
GO:0070160
|
tight junction
|
0.046906037145282
|
131
|
65
|
3
|
0.0461538461538462
|
0.0229007633587786
|
|
GO:MF
|
GO:0001216
|
DNA-binding transcription activator activity
|
4.954802719032e-11
|
455
|
76
|
18
|
0.236842105263158
|
0.0395604395604396
|
|
GO:MF
|
GO:0001228
|
DNA-binding transcription activator activity, RNA polymerase II-specific
|
4.954802719032e-11
|
451
|
76
|
18
|
0.236842105263158
|
0.0399113082039911
|
|
GO:MF
|
GO:0008134
|
transcription factor binding
|
1.6192940891757e-07
|
676
|
95
|
19
|
0.2
|
0.0281065088757396
|
|
GO:MF
|
GO:0061629
|
RNA polymerase II-specific DNA-binding transcription factor binding
|
3.80096370540443e-07
|
281
|
120
|
14
|
0.116666666666667
|
0.0498220640569395
|
|
GO:MF
|
GO:0140297
|
DNA-binding transcription factor binding
|
3.80096370540443e-07
|
391
|
120
|
16
|
0.133333333333333
|
0.040920716112532
|
|
GO:MF
|
GO:0000978
|
RNA polymerase II cis-regulatory region sequence-specific DNA binding
|
1.03066045540208e-06
|
1190
|
124
|
27
|
0.217741935483871
|
0.0226890756302521
|
|
GO:MF
|
GO:0140110
|
transcription regulator activity
|
1.21208297526187e-06
|
1935
|
124
|
35
|
0.282258064516129
|
0.0180878552971576
|
|
GO:MF
|
GO:0000987
|
cis-regulatory region sequence-specific DNA binding
|
1.21208297526187e-06
|
1209
|
124
|
27
|
0.217741935483871
|
0.0223325062034739
|
|
GO:MF
|
GO:0043565
|
sequence-specific DNA binding
|
3.2818634141031e-06
|
1673
|
124
|
31
|
0.25
|
0.0185295875672445
|
|
GO:MF
|
GO:1990837
|
sequence-specific double-stranded DNA binding
|
3.2818634141031e-06
|
1570
|
124
|
30
|
0.241935483870968
|
0.0191082802547771
|
|
GO:MF
|
GO:0019899
|
enzyme binding
|
3.2818634141031e-06
|
2074
|
96
|
30
|
0.3125
|
0.0144648023143684
|
|
GO:MF
|
GO:0000981
|
DNA-binding transcription factor activity, RNA polymerase II-specific
|
3.2818634141031e-06
|
1375
|
8
|
7
|
0.875
|
0.00509090909090909
|
|
GO:MF
|
GO:0000977
|
RNA polymerase II transcription regulatory region sequence-specific DNA binding
|
3.2818634141031e-06
|
1408
|
8
|
7
|
0.875
|
0.00497159090909091
|
|
GO:MF
|
GO:0003677
|
DNA binding
|
3.2818634141031e-06
|
2486
|
12
|
10
|
0.833333333333333
|
0.00402252614641995
|
|
GO:MF
|
GO:0003700
|
DNA-binding transcription factor activity
|
3.53994710214413e-06
|
1441
|
8
|
7
|
0.875
|
0.00485773768216516
|
|
GO:MF
|
GO:0000976
|
transcription regulatory region sequence-specific DNA binding
|
3.59556092876172e-06
|
1507
|
124
|
29
|
0.233870967741935
|
0.0192435301924353
|
|
GO:MF
|
GO:0001067
|
regulatory region nucleic acid binding
|
3.59556092876172e-06
|
1509
|
124
|
29
|
0.233870967741935
|
0.0192180251822399
|
|
GO:MF
|
GO:0035925
|
mRNA 3'-UTR AU-rich region binding
|
6.6454797485046e-06
|
25
|
42
|
4
|
0.0952380952380952
|
0.16
|
|
GO:MF
|
GO:0003690
|
double-stranded DNA binding
|
7.43313955840355e-06
|
1664
|
124
|
30
|
0.241935483870968
|
0.0180288461538462
|
|
GO:MF
|
GO:0005515
|
protein binding
|
4.48261063016705e-05
|
14767
|
98
|
94
|
0.959183673469388
|
0.00636554479582854
|
|
GO:MF
|
GO:0019900
|
kinase binding
|
0.000132557487695158
|
752
|
120
|
17
|
0.141666666666667
|
0.0226063829787234
|
|
GO:MF
|
GO:0003676
|
nucleic acid binding
|
0.000151292499600114
|
4276
|
8
|
8
|
1
|
0.00187090739008419
|
|
GO:MF
|
GO:0019901
|
protein kinase binding
|
0.000503216659155421
|
673
|
120
|
15
|
0.125
|
0.0222882615156018
|
|
GO:MF
|
GO:0019207
|
kinase regulator activity
|
0.000583448704087636
|
221
|
104
|
8
|
0.0769230769230769
|
0.0361990950226244
|
|
GO:MF
|
GO:0004860
|
protein kinase inhibitor activity
|
0.000752065133694781
|
70
|
104
|
5
|
0.0480769230769231
|
0.0714285714285714
|
|
GO:MF
|
GO:0035257
|
nuclear hormone receptor binding
|
0.000758977289626396
|
148
|
120
|
7
|
0.0583333333333333
|
0.0472972972972973
|
|
GO:MF
|
GO:0098772
|
molecular function regulator
|
0.000758977289626396
|
2363
|
118
|
31
|
0.26271186440678
|
0.0131189166314008
|
|
GO:MF
|
GO:0019210
|
kinase inhibitor activity
|
0.000881760081029367
|
74
|
104
|
5
|
0.0480769230769231
|
0.0675675675675676
|
|
GO:MF
|
GO:0031625
|
ubiquitin protein ligase binding
|
0.000894523016943951
|
296
|
85
|
8
|
0.0941176470588235
|
0.027027027027027
|
|
GO:MF
|
GO:0001102
|
RNA polymerase II activating transcription factor binding
|
0.00118509305299015
|
49
|
88
|
4
|
0.0454545454545455
|
0.0816326530612245
|
|
GO:MF
|
GO:0044389
|
ubiquitin-like protein ligase binding
|
0.00122747202211224
|
314
|
85
|
8
|
0.0941176470588235
|
0.0254777070063694
|
|
GO:MF
|
GO:0035259
|
glucocorticoid receptor binding
|
0.00122747202211224
|
14
|
120
|
3
|
0.025
|
0.214285714285714
|
|
GO:MF
|
GO:0019887
|
protein kinase regulator activity
|
0.00125730289658389
|
191
|
104
|
7
|
0.0673076923076923
|
0.0366492146596859
|
|
GO:MF
|
GO:0003714
|
transcription corepressor activity
|
0.0017133386604434
|
187
|
113
|
7
|
0.0619469026548673
|
0.0374331550802139
|
|
GO:MF
|
GO:0035014
|
phosphatidylinositol 3-kinase regulator activity
|
0.0017133386604434
|
20
|
96
|
3
|
0.03125
|
0.15
|
|
GO:MF
|
GO:1901363
|
heterocyclic compound binding
|
0.00177760779361799
|
6228
|
8
|
8
|
1
|
0.00128452151573539
|
|
GO:MF
|
GO:0003682
|
chromatin binding
|
0.00180313570253011
|
571
|
80
|
10
|
0.125
|
0.0175131348511384
|
|
GO:MF
|
GO:0097159
|
organic cyclic compound binding
|
0.00187275095404909
|
6309
|
8
|
8
|
1
|
0.00126802979870027
|
|
GO:MF
|
GO:0051427
|
hormone receptor binding
|
0.00189339388950965
|
182
|
120
|
7
|
0.0583333333333333
|
0.0384615384615385
|
|
GO:MF
|
GO:0051087
|
chaperone binding
|
0.00210417067066227
|
103
|
97
|
5
|
0.0515463917525773
|
0.0485436893203883
|
|
GO:MF
|
GO:0046332
|
SMAD binding
|
0.00217473457941267
|
78
|
28
|
3
|
0.107142857142857
|
0.0384615384615385
|
|
GO:MF
|
GO:0031072
|
heat shock protein binding
|
0.00271972451525786
|
127
|
85
|
5
|
0.0588235294117647
|
0.0393700787401575
|
|
GO:MF
|
GO:0001671
|
ATPase activator activity
|
0.00271972451525786
|
25
|
97
|
3
|
0.0309278350515464
|
0.12
|
|
GO:MF
|
GO:0051082
|
unfolded protein binding
|
0.00271972451525786
|
112
|
97
|
5
|
0.0515463917525773
|
0.0446428571428571
|
|
GO:MF
|
GO:1990841
|
promoter-specific chromatin binding
|
0.00364053434916469
|
58
|
113
|
4
|
0.0353982300884956
|
0.0689655172413793
|
|
GO:MF
|
GO:0071889
|
14-3-3 protein binding
|
0.00440342926789212
|
31
|
95
|
3
|
0.0315789473684211
|
0.0967741935483871
|
|
GO:MF
|
GO:0003730
|
mRNA 3'-UTR binding
|
0.00532413707609386
|
178
|
42
|
4
|
0.0952380952380952
|
0.0224719101123595
|
|
GO:MF
|
GO:0042826
|
histone deacetylase binding
|
0.00694131179271246
|
116
|
31
|
3
|
0.0967741935483871
|
0.0258620689655172
|
|
GO:MF
|
GO:0051019
|
mitogen-activated protein kinase binding
|
0.00695031497817472
|
32
|
113
|
3
|
0.0265486725663717
|
0.09375
|
|
GO:MF
|
GO:0060590
|
ATPase regulator activity
|
0.0103465943517348
|
44
|
97
|
3
|
0.0309278350515464
|
0.0681818181818182
|
|
GO:MF
|
GO:0016922
|
nuclear receptor binding
|
0.012601978464775
|
105
|
44
|
3
|
0.0681818181818182
|
0.0285714285714286
|
|
GO:MF
|
GO:0001664
|
G protein-coupled receptor binding
|
0.0175258334214298
|
292
|
90
|
6
|
0.0666666666666667
|
0.0205479452054795
|
|
GO:MF
|
GO:0015370
|
solute:sodium symporter activity
|
0.0175258334214298
|
72
|
74
|
3
|
0.0405405405405405
|
0.0416666666666667
|
|
GO:MF
|
GO:0046982
|
protein heterodimerization activity
|
0.0216427016730696
|
357
|
17
|
3
|
0.176470588235294
|
0.00840336134453781
|
|
GO:MF
|
GO:0098531
|
ligand-activated transcription factor activity
|
0.0263470903982954
|
54
|
120
|
3
|
0.025
|
0.0555555555555556
|
|
GO:MF
|
GO:0004879
|
nuclear receptor activity
|
0.0263470903982954
|
54
|
120
|
3
|
0.025
|
0.0555555555555556
|
|
GO:MF
|
GO:0005488
|
binding
|
0.0292775848503691
|
17074
|
81
|
80
|
0.987654320987654
|
0.00468548670493148
|
|
GO:MF
|
GO:0005125
|
cytokine activity
|
0.0296681826746755
|
235
|
58
|
4
|
0.0689655172413793
|
0.0170212765957447
|
|
GO:MF
|
GO:0015294
|
solute:cation symporter activity
|
0.033986721201726
|
99
|
74
|
3
|
0.0405405405405405
|
0.0303030303030303
|
|
GO:MF
|
GO:0030234
|
enzyme regulator activity
|
0.0406403914826851
|
1248
|
104
|
14
|
0.134615384615385
|
0.0112179487179487
|
|
GO:MF
|
GO:0004672
|
protein kinase activity
|
0.0424686246091158
|
587
|
114
|
9
|
0.0789473684210526
|
0.0153321976149915
|
|
GO:MF
|
GO:0005102
|
signaling receptor binding
|
0.0426385700939552
|
1672
|
120
|
19
|
0.158333333333333
|
0.0113636363636364
|
|
GO:MF
|
GO:0003727
|
single-stranded RNA binding
|
0.0438585213414927
|
92
|
92
|
3
|
0.0326086956521739
|
0.0326086956521739
|
|
GO:MF
|
GO:0008013
|
beta-catenin binding
|
0.0451315199163293
|
87
|
100
|
3
|
0.03
|
0.0344827586206897
|
|
GO:MF
|
GO:0003712
|
transcription coregulator activity
|
0.0455403875027341
|
506
|
113
|
8
|
0.0707964601769911
|
0.0158102766798419
|
|
GO:MF
|
GO:0042802
|
identical protein binding
|
0.0455403875027341
|
2067
|
41
|
10
|
0.24390243902439
|
0.00483792936623125
|
|
KEGG
|
KEGG:04668
|
TNF signaling pathway
|
4.45070745600905e-05
|
112
|
75
|
9
|
0.12
|
0.0803571428571429
|
|
KEGG
|
KEGG:04380
|
Osteoclast differentiation
|
4.45070745600905e-05
|
125
|
6
|
4
|
0.666666666666667
|
0.032
|
|
KEGG
|
KEGG:04657
|
IL-17 signaling pathway
|
4.45070745600905e-05
|
92
|
65
|
8
|
0.123076923076923
|
0.0869565217391304
|
|
KEGG
|
KEGG:05031
|
Amphetamine addiction
|
0.000464052881925177
|
69
|
6
|
3
|
0.5
|
0.0434782608695652
|
|
KEGG
|
KEGG:04010
|
MAPK signaling pathway
|
0.00254854516800913
|
294
|
30
|
7
|
0.233333333333333
|
0.0238095238095238
|
|
KEGG
|
KEGG:05120
|
Epithelial cell signaling in Helicobacter pylori infection
|
0.0036413159268234
|
70
|
58
|
5
|
0.0862068965517241
|
0.0714285714285714
|
|
KEGG
|
KEGG:04210
|
Apoptosis
|
0.00366269250506388
|
136
|
52
|
6
|
0.115384615384615
|
0.0441176470588235
|
|
KEGG
|
KEGG:05167
|
Kaposi sarcoma-associated herpesvirus infection
|
0.00366269250506388
|
193
|
37
|
6
|
0.162162162162162
|
0.0310880829015544
|
|
KEGG
|
KEGG:05166
|
Human T-cell leukemia virus 1 infection
|
0.00366269250506388
|
216
|
12
|
4
|
0.333333333333333
|
0.0185185185185185
|
|
KEGG
|
KEGG:04625
|
C-type lectin receptor signaling pathway
|
0.00366269250506388
|
104
|
68
|
6
|
0.0882352941176471
|
0.0576923076923077
|
|
KEGG
|
KEGG:04928
|
Parathyroid hormone synthesis, secretion and action
|
0.00374305702924407
|
106
|
44
|
5
|
0.113636363636364
|
0.0471698113207547
|
|
KEGG
|
KEGG:05418
|
Fluid shear stress and atherosclerosis
|
0.00501594417886745
|
138
|
9
|
3
|
0.333333333333333
|
0.0217391304347826
|
|
KEGG
|
KEGG:04915
|
Estrogen signaling pathway
|
0.00807953515952053
|
137
|
25
|
4
|
0.16
|
0.0291970802919708
|
|
KEGG
|
KEGG:05134
|
Legionellosis
|
0.00807953515952053
|
57
|
58
|
4
|
0.0689655172413793
|
0.0701754385964912
|
|
KEGG
|
KEGG:05210
|
Colorectal cancer
|
0.0116758689770155
|
86
|
78
|
5
|
0.0641025641025641
|
0.0581395348837209
|
|
KEGG
|
KEGG:05224
|
Breast cancer
|
0.0116758689770155
|
147
|
31
|
4
|
0.129032258064516
|
0.0272108843537415
|
|
KEGG
|
KEGG:04064
|
NF-kappa B signaling pathway
|
0.0135770076908332
|
102
|
78
|
5
|
0.0641025641025641
|
0.0490196078431373
|
|
KEGG
|
KEGG:05162
|
Measles
|
0.0135770076908332
|
139
|
35
|
4
|
0.114285714285714
|
0.0287769784172662
|
|
KEGG
|
KEGG:05130
|
Pathogenic Escherichia coli infection
|
0.0135770076908332
|
196
|
41
|
5
|
0.121951219512195
|
0.0255102040816327
|
|
KEGG
|
KEGG:04141
|
Protein processing in endoplasmic reticulum
|
0.0196823697532987
|
170
|
33
|
4
|
0.121212121212121
|
0.0235294117647059
|
|
KEGG
|
KEGG:05132
|
Salmonella infection
|
0.0247770286881458
|
249
|
13
|
3
|
0.230769230769231
|
0.0120481927710843
|
|
KEGG
|
KEGG:05160
|
Hepatitis C
|
0.0294365958778488
|
157
|
41
|
4
|
0.0975609756097561
|
0.0254777070063694
|
|
KEGG
|
KEGG:05169
|
Epstein-Barr virus infection
|
0.0342591430178927
|
198
|
35
|
4
|
0.114285714285714
|
0.0202020202020202
|
|
KEGG
|
KEGG:05145
|
Toxoplasmosis
|
0.0368670187505326
|
109
|
35
|
3
|
0.0857142857142857
|
0.0275229357798165
|
|
KEGG
|
KEGG:05222
|
Small cell lung cancer
|
0.0369178868353935
|
92
|
78
|
4
|
0.0512820512820513
|
0.0434782608695652
|
|
KEGG
|
KEGG:04925
|
Aldosterone synthesis and secretion
|
0.0468241506420058
|
98
|
44
|
3
|
0.0681818181818182
|
0.0306122448979592
|
|
MIRNA
|
MIRNA:hsa-miR-181c-5p
|
hsa-miR-181c-5p
|
1.26357773466754e-05
|
286
|
118
|
15
|
0.127118644067797
|
0.0524475524475524
|
|
MIRNA
|
MIRNA:hsa-miR-101-3p
|
hsa-miR-101-3p
|
1.26357773466754e-05
|
391
|
66
|
13
|
0.196969696969697
|
0.0332480818414322
|
|
MIRNA
|
MIRNA:hsa-miR-181b-5p
|
hsa-miR-181b-5p
|
2.24680257583309e-05
|
370
|
118
|
16
|
0.135593220338983
|
0.0432432432432432
|
|
MIRNA
|
MIRNA:hsa-miR-181a-5p
|
hsa-miR-181a-5p
|
5.25636844692053e-05
|
543
|
90
|
16
|
0.177777777777778
|
0.0294659300184162
|
|
MIRNA
|
MIRNA:hsa-miR-181d-5p
|
hsa-miR-181d-5p
|
5.25636844692053e-05
|
283
|
90
|
12
|
0.133333333333333
|
0.0424028268551237
|
|
MIRNA
|
MIRNA:hsa-miR-663a
|
hsa-miR-663a
|
0.000306374279684119
|
143
|
36
|
6
|
0.166666666666667
|
0.041958041958042
|
|
MIRNA
|
MIRNA:hsa-miR-93-5p
|
hsa-miR-93-5p
|
0.000306374279684119
|
1213
|
100
|
24
|
0.24
|
0.0197856553998351
|
|
MIRNA
|
MIRNA:hsa-miR-29a-3p
|
hsa-miR-29a-3p
|
0.000362901760957996
|
264
|
100
|
11
|
0.11
|
0.0416666666666667
|
|
MIRNA
|
MIRNA:hsa-miR-17-5p
|
hsa-miR-17-5p
|
0.000496853846916211
|
1174
|
100
|
23
|
0.23
|
0.0195911413969336
|
|
MIRNA
|
MIRNA:hsa-miR-29c-3p
|
hsa-miR-29c-3p
|
0.000646601090756917
|
253
|
113
|
11
|
0.0973451327433628
|
0.0434782608695652
|
|
MIRNA
|
MIRNA:hsa-miR-199a-5p
|
hsa-miR-199a-5p
|
0.000680049366004265
|
164
|
113
|
9
|
0.079646017699115
|
0.0548780487804878
|
|
MIRNA
|
MIRNA:hsa-miR-30a-5p
|
hsa-miR-30a-5p
|
0.000680049366004265
|
730
|
97
|
17
|
0.175257731958763
|
0.0232876712328767
|
|
MIRNA
|
MIRNA:hsa-miR-139-5p
|
hsa-miR-139-5p
|
0.000701546956939782
|
104
|
17
|
4
|
0.235294117647059
|
0.0384615384615385
|
|
MIRNA
|
MIRNA:hsa-miR-524-5p
|
hsa-miR-524-5p
|
0.00110103202577996
|
171
|
64
|
7
|
0.109375
|
0.0409356725146199
|
|
MIRNA
|
MIRNA:hsa-miR-4262
|
hsa-miR-4262
|
0.00118250478122571
|
134
|
83
|
7
|
0.0843373493975904
|
0.0522388059701493
|
|
MIRNA
|
MIRNA:hsa-miR-106a-5p
|
hsa-miR-106a-5p
|
0.00210692170357279
|
709
|
100
|
16
|
0.16
|
0.0225669957686883
|
|
MIRNA
|
MIRNA:hsa-miR-6083
|
hsa-miR-6083
|
0.00210692170357279
|
175
|
71
|
7
|
0.0985915492957746
|
0.04
|
|
MIRNA
|
MIRNA:hsa-miR-526b-3p
|
hsa-miR-526b-3p
|
0.00217940920345192
|
635
|
100
|
15
|
0.15
|
0.0236220472440945
|
|
MIRNA
|
MIRNA:hsa-miR-4678
|
hsa-miR-4678
|
0.00287403353295492
|
67
|
81
|
5
|
0.0617283950617284
|
0.0746268656716418
|
|
MIRNA
|
MIRNA:hsa-miR-6507-5p
|
hsa-miR-6507-5p
|
0.00287403353295492
|
132
|
21
|
4
|
0.19047619047619
|
0.0303030303030303
|
|
MIRNA
|
MIRNA:hsa-miR-548ah-5p
|
hsa-miR-548ah-5p
|
0.00288882234165956
|
318
|
95
|
10
|
0.105263157894737
|
0.0314465408805031
|
|
MIRNA
|
MIRNA:hsa-miR-3609
|
hsa-miR-3609
|
0.00288882234165956
|
319
|
95
|
10
|
0.105263157894737
|
0.0313479623824451
|
|
MIRNA
|
MIRNA:hsa-miR-3614-3p
|
hsa-miR-3614-3p
|
0.00288882234165956
|
65
|
85
|
5
|
0.0588235294117647
|
0.0769230769230769
|
|
MIRNA
|
MIRNA:hsa-miR-4709-5p
|
hsa-miR-4709-5p
|
0.00300568062736719
|
90
|
32
|
4
|
0.125
|
0.0444444444444444
|
|
MIRNA
|
MIRNA:hsa-miR-148b-5p
|
hsa-miR-148b-5p
|
0.00316125262030807
|
52
|
20
|
3
|
0.15
|
0.0576923076923077
|
|
MIRNA
|
MIRNA:hsa-miR-1910-5p
|
hsa-miR-1910-5p
|
0.00316125262030807
|
58
|
101
|
5
|
0.0495049504950495
|
0.0862068965517241
|
|
MIRNA
|
MIRNA:hsa-miR-4704-3p
|
hsa-miR-4704-3p
|
0.00326503060571826
|
64
|
47
|
4
|
0.0851063829787234
|
0.0625
|
|
MIRNA
|
MIRNA:hsa-miR-27b-3p
|
hsa-miR-27b-3p
|
0.00378463556392448
|
419
|
77
|
10
|
0.12987012987013
|
0.0238663484486874
|
|
MIRNA
|
MIRNA:hsa-miR-335-5p
|
hsa-miR-335-5p
|
0.00400325419476685
|
2571
|
90
|
31
|
0.344444444444444
|
0.0120575651497472
|
|
MIRNA
|
MIRNA:hsa-miR-130b-3p
|
hsa-miR-130b-3p
|
0.00400325419476685
|
571
|
95
|
13
|
0.136842105263158
|
0.0227670753064799
|
|
MIRNA
|
MIRNA:hsa-miR-27a-3p
|
hsa-miR-27a-3p
|
0.00400325419476685
|
427
|
77
|
10
|
0.12987012987013
|
0.0234192037470726
|
|
MIRNA
|
MIRNA:hsa-miR-520d-5p
|
hsa-miR-520d-5p
|
0.00414333639614632
|
164
|
64
|
6
|
0.09375
|
0.0365853658536585
|
|
MIRNA
|
MIRNA:hsa-miR-4787-5p
|
hsa-miR-4787-5p
|
0.00419574743734865
|
35
|
36
|
3
|
0.0833333333333333
|
0.0857142857142857
|
|
MIRNA
|
MIRNA:hsa-miR-20a-5p
|
hsa-miR-20a-5p
|
0.00419574743734865
|
1064
|
100
|
19
|
0.19
|
0.0178571428571429
|
|
MIRNA
|
MIRNA:hsa-miR-4796-3p
|
hsa-miR-4796-3p
|
0.00436450354533262
|
285
|
95
|
9
|
0.0947368421052632
|
0.0315789473684211
|
|
MIRNA
|
MIRNA:hsa-miR-98-5p
|
hsa-miR-98-5p
|
0.0043651247160441
|
778
|
63
|
12
|
0.19047619047619
|
0.0154241645244216
|
|
MIRNA
|
MIRNA:hsa-miR-148a-3p
|
hsa-miR-148a-3p
|
0.0043651247160441
|
212
|
100
|
8
|
0.08
|
0.0377358490566038
|
|
MIRNA
|
MIRNA:hsa-miR-15a-5p
|
hsa-miR-15a-5p
|
0.00453440433957467
|
717
|
24
|
7
|
0.291666666666667
|
0.0097629009762901
|
|
MIRNA
|
MIRNA:hsa-miR-519d-3p
|
hsa-miR-519d-3p
|
0.00453440433957467
|
900
|
100
|
17
|
0.17
|
0.0188888888888889
|
|
MIRNA
|
MIRNA:hsa-miR-7-2-3p
|
hsa-miR-7-2-3p
|
0.00487889124013132
|
167
|
97
|
7
|
0.0721649484536082
|
0.0419161676646707
|
|
MIRNA
|
MIRNA:hsa-miR-20b-5p
|
hsa-miR-20b-5p
|
0.00494353393488585
|
911
|
100
|
17
|
0.17
|
0.0186608122941822
|
|
MIRNA
|
MIRNA:hsa-miR-30d-5p
|
hsa-miR-30d-5p
|
0.00518927892923347
|
367
|
96
|
10
|
0.104166666666667
|
0.0272479564032698
|
|
MIRNA
|
MIRNA:hsa-miR-30c-5p
|
hsa-miR-30c-5p
|
0.00518927892923347
|
519
|
96
|
12
|
0.125
|
0.023121387283237
|
|
MIRNA
|
MIRNA:hsa-miR-5688
|
hsa-miR-5688
|
0.00518927892923347
|
168
|
69
|
6
|
0.0869565217391304
|
0.0357142857142857
|
|
MIRNA
|
MIRNA:hsa-miR-4797-5p
|
hsa-miR-4797-5p
|
0.00543225335831254
|
212
|
106
|
8
|
0.0754716981132075
|
0.0377358490566038
|
|
MIRNA
|
MIRNA:hsa-miR-340-5p
|
hsa-miR-340-5p
|
0.00587028732057722
|
340
|
86
|
9
|
0.104651162790698
|
0.0264705882352941
|
|
MIRNA
|
MIRNA:hsa-miR-7-1-3p
|
hsa-miR-7-1-3p
|
0.00660335002315205
|
180
|
97
|
7
|
0.0721649484536082
|
0.0388888888888889
|
|
MIRNA
|
MIRNA:hsa-miR-199b-5p
|
hsa-miR-199b-5p
|
0.00660335002315205
|
109
|
113
|
6
|
0.0530973451327434
|
0.055045871559633
|
|
MIRNA
|
MIRNA:hsa-miR-627-5p
|
hsa-miR-627-5p
|
0.00660335002315205
|
69
|
24
|
3
|
0.125
|
0.0434782608695652
|
|
MIRNA
|
MIRNA:hsa-miR-495-3p
|
hsa-miR-495-3p
|
0.00701412681228457
|
182
|
69
|
6
|
0.0869565217391304
|
0.032967032967033
|
|
MIRNA
|
MIRNA:hsa-miR-570-3p
|
hsa-miR-570-3p
|
0.00735364231669206
|
178
|
101
|
7
|
0.0693069306930693
|
0.0393258426966292
|
|
MIRNA
|
MIRNA:hsa-miR-130a-3p
|
hsa-miR-130a-3p
|
0.00778987772253606
|
398
|
95
|
10
|
0.105263157894737
|
0.0251256281407035
|
|
MIRNA
|
MIRNA:hsa-miR-1284
|
hsa-miR-1284
|
0.00809781520747139
|
47
|
97
|
4
|
0.0412371134020619
|
0.0851063829787234
|
|
MIRNA
|
MIRNA:hsa-miR-624-3p
|
hsa-miR-624-3p
|
0.0081312572408424
|
101
|
83
|
5
|
0.0602409638554217
|
0.0495049504950495
|
|
MIRNA
|
MIRNA:hsa-miR-144-3p
|
hsa-miR-144-3p
|
0.00837823996239143
|
207
|
23
|
4
|
0.173913043478261
|
0.0193236714975845
|
|
MIRNA
|
MIRNA:hsa-let-7f-1-3p
|
hsa-let-7f-1-3p
|
0.00882501585810944
|
114
|
76
|
5
|
0.0657894736842105
|
0.043859649122807
|
|
MIRNA
|
MIRNA:hsa-miR-98-3p
|
hsa-miR-98-3p
|
0.00882501585810944
|
114
|
76
|
5
|
0.0657894736842105
|
0.043859649122807
|
|
MIRNA
|
MIRNA:hsa-miR-641
|
hsa-miR-641
|
0.00882501585810944
|
103
|
84
|
5
|
0.0595238095238095
|
0.0485436893203883
|
|
MIRNA
|
MIRNA:hsa-miR-937-5p
|
hsa-miR-937-5p
|
0.0089676481045096
|
83
|
24
|
3
|
0.125
|
0.036144578313253
|
|
MIRNA
|
MIRNA:hsa-miR-106b-5p
|
hsa-miR-106b-5p
|
0.0089676481045096
|
1084
|
100
|
18
|
0.18
|
0.0166051660516605
|
|
MIRNA
|
MIRNA:hsa-miR-4303
|
hsa-miR-4303
|
0.0089676481045096
|
100
|
20
|
3
|
0.15
|
0.03
|
|
MIRNA
|
MIRNA:hsa-let-7b-3p
|
hsa-let-7b-3p
|
0.0089676481045096
|
116
|
76
|
5
|
0.0657894736842105
|
0.0431034482758621
|
|
MIRNA
|
MIRNA:hsa-miR-421
|
hsa-miR-421
|
0.00934505595845645
|
235
|
83
|
7
|
0.0843373493975904
|
0.0297872340425532
|
|
MIRNA
|
MIRNA:hsa-miR-759
|
hsa-miR-759
|
0.00937986007882811
|
184
|
106
|
7
|
0.0660377358490566
|
0.0380434782608696
|
|
MIRNA
|
MIRNA:hsa-miR-29b-3p
|
hsa-miR-29b-3p
|
0.00968351584764764
|
260
|
100
|
8
|
0.08
|
0.0307692307692308
|
|
MIRNA
|
MIRNA:hsa-let-7f-2-3p
|
hsa-let-7f-2-3p
|
0.00979460426540586
|
98
|
52
|
4
|
0.0769230769230769
|
0.0408163265306122
|
|
MIRNA
|
MIRNA:hsa-miR-200c-3p
|
hsa-miR-200c-3p
|
0.00985007809874113
|
214
|
122
|
8
|
0.0655737704918033
|
0.0373831775700935
|
|
MIRNA
|
MIRNA:hsa-let-7a-3p
|
hsa-let-7a-3p
|
0.0104443631377856
|
123
|
76
|
5
|
0.0657894736842105
|
0.040650406504065
|
|
MIRNA
|
MIRNA:hsa-miR-296-3p
|
hsa-miR-296-3p
|
0.0104670547064479
|
255
|
57
|
6
|
0.105263157894737
|
0.0235294117647059
|
|
MIRNA
|
MIRNA:hsa-miR-8081
|
hsa-miR-8081
|
0.0110338706061883
|
94
|
24
|
3
|
0.125
|
0.0319148936170213
|
|
MIRNA
|
MIRNA:hsa-miR-124-3p
|
hsa-miR-124-3p
|
0.0113836694427779
|
1442
|
120
|
24
|
0.2
|
0.0166435506241331
|
|
MIRNA
|
MIRNA:hsa-miR-4699-3p
|
hsa-miR-4699-3p
|
0.0113839826900587
|
89
|
61
|
4
|
0.0655737704918033
|
0.0449438202247191
|
|
MIRNA
|
MIRNA:hsa-miR-221-3p
|
hsa-miR-221-3p
|
0.0113839826900587
|
366
|
113
|
10
|
0.0884955752212389
|
0.0273224043715847
|
|
MIRNA
|
MIRNA:hsa-miR-4487
|
hsa-miR-4487
|
0.0115482640414274
|
219
|
45
|
5
|
0.111111111111111
|
0.0228310502283105
|
|
MIRNA
|
MIRNA:hsa-miR-3666
|
hsa-miR-3666
|
0.0119302552553488
|
364
|
95
|
9
|
0.0947368421052632
|
0.0247252747252747
|
|
MIRNA
|
MIRNA:hsa-miR-4295
|
hsa-miR-4295
|
0.0122584022488194
|
366
|
95
|
9
|
0.0947368421052632
|
0.0245901639344262
|
|
MIRNA
|
MIRNA:hsa-miR-212-3p
|
hsa-miR-212-3p
|
0.0124890495146503
|
93
|
61
|
4
|
0.0655737704918033
|
0.043010752688172
|
|
MIRNA
|
MIRNA:hsa-miR-6845-3p
|
hsa-miR-6845-3p
|
0.013593056225423
|
262
|
40
|
5
|
0.125
|
0.0190839694656489
|
|
MIRNA
|
MIRNA:hsa-miR-140-3p
|
hsa-miR-140-3p
|
0.013593056225423
|
221
|
71
|
6
|
0.0845070422535211
|
0.0271493212669683
|
|
MIRNA
|
MIRNA:hsa-miR-301b-3p
|
hsa-miR-301b-3p
|
0.013593056225423
|
375
|
95
|
9
|
0.0947368421052632
|
0.024
|
|
MIRNA
|
MIRNA:hsa-miR-6126
|
hsa-miR-6126
|
0.013593056225423
|
70
|
84
|
4
|
0.0476190476190476
|
0.0571428571428571
|
|
MIRNA
|
MIRNA:hsa-miR-152-3p
|
hsa-miR-152-3p
|
0.0136009841953615
|
157
|
100
|
6
|
0.06
|
0.0382165605095541
|
|
MIRNA
|
MIRNA:hsa-miR-30e-3p
|
hsa-miR-30e-3p
|
0.0141330879026077
|
173
|
35
|
4
|
0.114285714285714
|
0.023121387283237
|
|
MIRNA
|
MIRNA:hsa-miR-1185-1-3p
|
hsa-miR-1185-1-3p
|
0.0142573710952755
|
116
|
52
|
4
|
0.0769230769230769
|
0.0344827586206897
|
|
MIRNA
|
MIRNA:hsa-miR-4695-5p
|
hsa-miR-4695-5p
|
0.015166813561733
|
366
|
45
|
6
|
0.133333333333333
|
0.0163934426229508
|
|
MIRNA
|
MIRNA:hsa-miR-1185-2-3p
|
hsa-miR-1185-2-3p
|
0.0151890439463053
|
119
|
52
|
4
|
0.0769230769230769
|
0.0336134453781513
|
|
MIRNA
|
MIRNA:hsa-miR-335-3p
|
hsa-miR-335-3p
|
0.0172205836528562
|
302
|
99
|
8
|
0.0808080808080808
|
0.0264900662251656
|
|
MIRNA
|
MIRNA:hsa-miR-301a-3p
|
hsa-miR-301a-3p
|
0.017601199059185
|
395
|
95
|
9
|
0.0947368421052632
|
0.0227848101265823
|
|
MIRNA
|
MIRNA:hsa-miR-92a-3p
|
hsa-miR-92a-3p
|
0.017601199059185
|
1404
|
100
|
20
|
0.2
|
0.0142450142450142
|
|
MIRNA
|
MIRNA:hsa-miR-454-3p
|
hsa-miR-454-3p
|
0.017601199059185
|
396
|
95
|
9
|
0.0947368421052632
|
0.0227272727272727
|
|
MIRNA
|
MIRNA:hsa-miR-506-3p
|
hsa-miR-506-3p
|
0.0183683321075469
|
128
|
89
|
5
|
0.0561797752808989
|
0.0390625
|
|
MIRNA
|
MIRNA:hsa-miR-19b-3p
|
hsa-miR-19b-3p
|
0.018811389308419
|
712
|
101
|
13
|
0.128712871287129
|
0.0182584269662921
|
|
MIRNA
|
MIRNA:hsa-let-7b-5p
|
hsa-let-7b-5p
|
0.0189969323268484
|
1212
|
61
|
13
|
0.213114754098361
|
0.0107260726072607
|
|
MIRNA
|
MIRNA:hsa-miR-223-3p
|
hsa-miR-223-3p
|
0.0191401796729138
|
98
|
69
|
4
|
0.0579710144927536
|
0.0408163265306122
|
|
MIRNA
|
MIRNA:hsa-miR-411-3p
|
hsa-miR-411-3p
|
0.0202575165161115
|
72
|
96
|
4
|
0.0416666666666667
|
0.0555555555555556
|
|
MIRNA
|
MIRNA:hsa-miR-543
|
hsa-miR-543
|
0.0204678280399272
|
105
|
113
|
5
|
0.0442477876106195
|
0.0476190476190476
|
|
MIRNA
|
MIRNA:hsa-miR-6077
|
hsa-miR-6077
|
0.0206656889020194
|
278
|
12
|
3
|
0.25
|
0.0107913669064748
|
|
MIRNA
|
MIRNA:hsa-miR-4676-5p
|
hsa-miR-4676-5p
|
0.0206656889020194
|
128
|
55
|
4
|
0.0727272727272727
|
0.03125
|
|
MIRNA
|
MIRNA:hsa-miR-575
|
hsa-miR-575
|
0.0206656889020194
|
128
|
55
|
4
|
0.0727272727272727
|
0.03125
|
|
MIRNA
|
MIRNA:hsa-miR-4495
|
hsa-miR-4495
|
0.0213172324624145
|
126
|
96
|
5
|
0.0520833333333333
|
0.0396825396825397
|
|
MIRNA
|
MIRNA:hsa-miR-4493
|
hsa-miR-4493
|
0.022148138761588
|
77
|
94
|
4
|
0.0425531914893617
|
0.051948051948052
|
|
MIRNA
|
MIRNA:hsa-miR-30b-5p
|
hsa-miR-30b-5p
|
0.022148138761588
|
414
|
96
|
9
|
0.09375
|
0.0217391304347826
|
|
MIRNA
|
MIRNA:hsa-miR-605-5p
|
hsa-miR-605-5p
|
0.022148138761588
|
111
|
65
|
4
|
0.0615384615384615
|
0.036036036036036
|
|
MIRNA
|
MIRNA:hsa-miR-5680
|
hsa-miR-5680
|
0.0222033365706565
|
188
|
39
|
4
|
0.102564102564103
|
0.0212765957446809
|
|
MIRNA
|
MIRNA:hsa-miR-380-3p
|
hsa-miR-380-3p
|
0.0224242796405466
|
76
|
96
|
4
|
0.0416666666666667
|
0.0526315789473684
|
|
MIRNA
|
MIRNA:hsa-miR-6508-5p
|
hsa-miR-6508-5p
|
0.0238968368164739
|
74
|
101
|
4
|
0.0396039603960396
|
0.0540540540540541
|
|
MIRNA
|
MIRNA:hsa-miR-518a-5p
|
hsa-miR-518a-5p
|
0.024051642412316
|
158
|
118
|
6
|
0.0508474576271186
|
0.0379746835443038
|
|
MIRNA
|
MIRNA:hsa-miR-19a-3p
|
hsa-miR-19a-3p
|
0.0241239687061102
|
569
|
101
|
11
|
0.108910891089109
|
0.0193321616871705
|
|
MIRNA
|
MIRNA:hsa-miR-8067
|
hsa-miR-8067
|
0.0241239687061102
|
75
|
101
|
4
|
0.0396039603960396
|
0.0533333333333333
|
|
MIRNA
|
MIRNA:hsa-miR-4803
|
hsa-miR-4803
|
0.0241239687061102
|
93
|
81
|
4
|
0.0493827160493827
|
0.043010752688172
|
|
MIRNA
|
MIRNA:hsa-miR-30e-5p
|
hsa-miR-30e-5p
|
0.0251933528513171
|
346
|
96
|
8
|
0.0833333333333333
|
0.023121387283237
|
|
MIRNA
|
MIRNA:hsa-miR-200b-3p
|
hsa-miR-200b-3p
|
0.0263317824168368
|
185
|
71
|
5
|
0.0704225352112676
|
0.027027027027027
|
|
MIRNA
|
MIRNA:hsa-miR-548n
|
hsa-miR-548n
|
0.0265745158440823
|
259
|
101
|
7
|
0.0693069306930693
|
0.027027027027027
|
|
MIRNA
|
MIRNA:hsa-miR-132-3p
|
hsa-miR-132-3p
|
0.027205763000582
|
252
|
32
|
4
|
0.125
|
0.0158730158730159
|
|
MIRNA
|
MIRNA:hsa-miR-4659b-3p
|
hsa-miR-4659b-3p
|
0.027205763000582
|
228
|
86
|
6
|
0.0697674418604651
|
0.0263157894736842
|
|
MIRNA
|
MIRNA:hsa-miR-379-3p
|
hsa-miR-379-3p
|
0.027205763000582
|
83
|
96
|
4
|
0.0416666666666667
|
0.0481927710843374
|
|
MIRNA
|
MIRNA:hsa-miR-505-5p
|
hsa-miR-505-5p
|
0.027205763000582
|
133
|
100
|
5
|
0.05
|
0.037593984962406
|
|
MIRNA
|
MIRNA:hsa-miR-4761-3p
|
hsa-miR-4761-3p
|
0.027205763000582
|
43
|
88
|
3
|
0.0340909090909091
|
0.0697674418604651
|
|
MIRNA
|
MIRNA:hsa-miR-4659a-3p
|
hsa-miR-4659a-3p
|
0.027205763000582
|
228
|
86
|
6
|
0.0697674418604651
|
0.0263157894736842
|
|
MIRNA
|
MIRNA:hsa-miR-6073
|
hsa-miR-6073
|
0.0275039834219507
|
95
|
85
|
4
|
0.0470588235294118
|
0.0421052631578947
|
|
MIRNA
|
MIRNA:hsa-miR-8066
|
hsa-miR-8066
|
0.0280412103011582
|
98
|
83
|
4
|
0.0481927710843374
|
0.0408163265306122
|
|
MIRNA
|
MIRNA:hsa-miR-4317
|
hsa-miR-4317
|
0.0288496589776876
|
47
|
85
|
3
|
0.0352941176470588
|
0.0638297872340425
|
|
MIRNA
|
MIRNA:hsa-miR-1908-5p
|
hsa-miR-1908-5p
|
0.0288496589776876
|
134
|
62
|
4
|
0.0645161290322581
|
0.0298507462686567
|
|
MIRNA
|
MIRNA:hsa-let-7c-5p
|
hsa-let-7c-5p
|
0.0288496589776876
|
514
|
54
|
7
|
0.12962962962963
|
0.0136186770428016
|
|
MIRNA
|
MIRNA:hsa-miR-3173-3p
|
hsa-miR-3173-3p
|
0.0288496589776876
|
134
|
104
|
5
|
0.0480769230769231
|
0.0373134328358209
|
|
MIRNA
|
MIRNA:hsa-miR-25-3p
|
hsa-miR-25-3p
|
0.0288496589776876
|
515
|
28
|
5
|
0.178571428571429
|
0.00970873786407767
|
|
MIRNA
|
MIRNA:hsa-miR-4515
|
hsa-miR-4515
|
0.0288496589776876
|
40
|
100
|
3
|
0.03
|
0.075
|
|
MIRNA
|
MIRNA:hsa-miR-4423-5p
|
hsa-miR-4423-5p
|
0.0288496589776876
|
209
|
97
|
6
|
0.0618556701030928
|
0.0287081339712919
|
|
MIRNA
|
MIRNA:hsa-miR-3914
|
hsa-miR-3914
|
0.0288496589776876
|
56
|
71
|
3
|
0.0422535211267606
|
0.0535714285714286
|
|
MIRNA
|
MIRNA:hsa-miR-194-3p
|
hsa-miR-194-3p
|
0.0288496589776876
|
131
|
64
|
4
|
0.0625
|
0.0305343511450382
|
|
MIRNA
|
MIRNA:hsa-miR-377-3p
|
hsa-miR-377-3p
|
0.0289242418683069
|
266
|
53
|
5
|
0.0943396226415094
|
0.018796992481203
|
|
MIRNA
|
MIRNA:hsa-miR-206
|
hsa-miR-206
|
0.0289242418683069
|
121
|
70
|
4
|
0.0571428571428571
|
0.0330578512396694
|
|
MIRNA
|
MIRNA:hsa-miR-3617-5p
|
hsa-miR-3617-5p
|
0.0290137149741566
|
101
|
84
|
4
|
0.0476190476190476
|
0.0396039603960396
|
|
MIRNA
|
MIRNA:hsa-miR-6749-3p
|
hsa-miR-6749-3p
|
0.0299575210251553
|
385
|
23
|
4
|
0.173913043478261
|
0.0103896103896104
|
|
MIRNA
|
MIRNA:hsa-miR-1305
|
hsa-miR-1305
|
0.0300108899856678
|
194
|
45
|
4
|
0.0888888888888889
|
0.0206185567010309
|
|
MIRNA
|
MIRNA:hsa-miR-3529-5p
|
hsa-miR-3529-5p
|
0.0300108899856678
|
83
|
104
|
4
|
0.0384615384615385
|
0.0481927710843374
|
|
MIRNA
|
MIRNA:hsa-miR-6891-5p
|
hsa-miR-6891-5p
|
0.0300108899856678
|
137
|
104
|
5
|
0.0480769230769231
|
0.0364963503649635
|
|
MIRNA
|
MIRNA:hsa-miR-495-5p
|
hsa-miR-495-5p
|
0.0300108899856678
|
92
|
45
|
3
|
0.0666666666666667
|
0.0326086956521739
|
|
MIRNA
|
MIRNA:hsa-let-7d-5p
|
hsa-let-7d-5p
|
0.0307476915253059
|
391
|
54
|
6
|
0.111111111111111
|
0.0153452685421995
|
|
MIRNA
|
MIRNA:hsa-miR-3160-3p
|
hsa-miR-3160-3p
|
0.0307720978561663
|
196
|
45
|
4
|
0.0888888888888889
|
0.0204081632653061
|
|
MIRNA
|
MIRNA:hsa-miR-379-5p
|
hsa-miR-379-5p
|
0.0315061848775183
|
85
|
104
|
4
|
0.0384615384615385
|
0.0470588235294118
|
|
MIRNA
|
MIRNA:hsa-miR-6721-5p
|
hsa-miR-6721-5p
|
0.0325922722682835
|
234
|
63
|
5
|
0.0793650793650794
|
0.0213675213675214
|
|
MIRNA
|
MIRNA:hsa-miR-193a-3p
|
hsa-miR-193a-3p
|
0.0325922722682835
|
128
|
70
|
4
|
0.0571428571428571
|
0.03125
|
|
MIRNA
|
MIRNA:hsa-miR-3144-5p
|
hsa-miR-3144-5p
|
0.0338182807960778
|
70
|
63
|
3
|
0.0476190476190476
|
0.0428571428571429
|
|
MIRNA
|
MIRNA:hsa-miR-4270
|
hsa-miR-4270
|
0.0343765578247172
|
216
|
21
|
3
|
0.142857142857143
|
0.0138888888888889
|
|
MIRNA
|
MIRNA:hsa-miR-4801
|
hsa-miR-4801
|
0.0345374052077969
|
97
|
95
|
4
|
0.0421052631578947
|
0.0412371134020619
|
|
MIRNA
|
MIRNA:hsa-miR-1277-5p
|
hsa-miR-1277-5p
|
0.0345374052077969
|
621
|
36
|
6
|
0.166666666666667
|
0.00966183574879227
|
|
MIRNA
|
MIRNA:hsa-miR-6754-5p
|
hsa-miR-6754-5p
|
0.0345374052077969
|
217
|
21
|
3
|
0.142857142857143
|
0.0138248847926267
|
|
MIRNA
|
MIRNA:hsa-miR-4441
|
hsa-miR-4441
|
0.0345374052077969
|
218
|
21
|
3
|
0.142857142857143
|
0.0137614678899083
|
|
MIRNA
|
MIRNA:hsa-miR-6787-5p
|
hsa-miR-6787-5p
|
0.0345374052077969
|
126
|
36
|
3
|
0.0833333333333333
|
0.0238095238095238
|
|
MIRNA
|
MIRNA:hsa-miR-3163
|
hsa-miR-3163
|
0.0345374052077969
|
338
|
86
|
7
|
0.0813953488372093
|
0.0207100591715976
|
|
MIRNA
|
MIRNA:hsa-miR-4731-3p
|
hsa-miR-4731-3p
|
0.034923021786131
|
98
|
95
|
4
|
0.0421052631578947
|
0.0408163265306122
|
|
MIRNA
|
MIRNA:hsa-miR-3611
|
hsa-miR-3611
|
0.034923021786131
|
99
|
94
|
4
|
0.0425531914893617
|
0.0404040404040404
|
|
MIRNA
|
MIRNA:hsa-miR-155-5p
|
hsa-miR-155-5p
|
0.034923021786131
|
900
|
18
|
5
|
0.277777777777778
|
0.00555555555555556
|
|
MIRNA
|
MIRNA:hsa-let-7a-5p
|
hsa-let-7a-5p
|
0.0357666355234026
|
637
|
100
|
11
|
0.11
|
0.0172684458398744
|
|
MIRNA
|
MIRNA:hsa-miR-6837-5p
|
hsa-miR-6837-5p
|
0.0358516177419716
|
123
|
38
|
3
|
0.0789473684210526
|
0.024390243902439
|
|
MIRNA
|
MIRNA:hsa-miR-4763-3p
|
hsa-miR-4763-3p
|
0.0358516177419716
|
282
|
17
|
3
|
0.176470588235294
|
0.0106382978723404
|
|
MIRNA
|
MIRNA:hsa-miR-410-3p
|
hsa-miR-410-3p
|
0.0358516177419716
|
248
|
89
|
6
|
0.0674157303370786
|
0.0241935483870968
|
|
MIRNA
|
MIRNA:hsa-miR-1207-5p
|
hsa-miR-1207-5p
|
0.0361868901092931
|
287
|
17
|
3
|
0.176470588235294
|
0.0104529616724739
|
|
MIRNA
|
MIRNA:hsa-miR-378a-3p
|
hsa-miR-378a-3p
|
0.0361868901092931
|
165
|
94
|
5
|
0.0531914893617021
|
0.0303030303030303
|
|
MIRNA
|
MIRNA:hsa-miR-4685-5p
|
hsa-miR-4685-5p
|
0.0361868901092931
|
124
|
38
|
3
|
0.0789473684210526
|
0.0241935483870968
|
|
MIRNA
|
MIRNA:hsa-miR-216a-3p
|
hsa-miR-216a-3p
|
0.0361868901092931
|
243
|
64
|
5
|
0.078125
|
0.0205761316872428
|
|
MIRNA
|
MIRNA:hsa-miR-3681-3p
|
hsa-miR-3681-3p
|
0.0361868901092931
|
243
|
64
|
5
|
0.078125
|
0.0205761316872428
|
|
MIRNA
|
MIRNA:hsa-miR-26a-5p
|
hsa-miR-26a-5p
|
0.0361868901092931
|
457
|
83
|
8
|
0.0963855421686747
|
0.0175054704595186
|
|
MIRNA
|
MIRNA:hsa-miR-30a-3p
|
hsa-miR-30a-3p
|
0.0364116199086107
|
193
|
25
|
3
|
0.12
|
0.0155440414507772
|
|
MIRNA
|
MIRNA:hsa-miR-655-5p
|
hsa-miR-655-5p
|
0.0380401318466359
|
122
|
40
|
3
|
0.075
|
0.0245901639344262
|
|
MIRNA
|
MIRNA:hsa-miR-520f-3p
|
hsa-miR-520f-3p
|
0.0380401318466359
|
103
|
95
|
4
|
0.0421052631578947
|
0.0388349514563107
|
|
MIRNA
|
MIRNA:hsa-miR-744-3p
|
hsa-miR-744-3p
|
0.0385959704232009
|
235
|
97
|
6
|
0.0618556701030928
|
0.025531914893617
|
|
MIRNA
|
MIRNA:hsa-miR-331-5p
|
hsa-miR-331-5p
|
0.0385959704232009
|
63
|
78
|
3
|
0.0384615384615385
|
0.0476190476190476
|
|
MIRNA
|
MIRNA:hsa-miR-455-3p
|
hsa-miR-455-3p
|
0.0385959704232009
|
782
|
61
|
9
|
0.147540983606557
|
0.0115089514066496
|
|
MIRNA
|
MIRNA:hsa-miR-1296-3p
|
hsa-miR-1296-3p
|
0.0388957103890384
|
99
|
100
|
4
|
0.04
|
0.0404040404040404
|
|
MIRNA
|
MIRNA:hsa-miR-449a
|
hsa-miR-449a
|
0.0389861638611952
|
158
|
63
|
4
|
0.0634920634920635
|
0.0253164556962025
|
|
MIRNA
|
MIRNA:hsa-miR-568
|
hsa-miR-568
|
0.0389861638611952
|
74
|
67
|
3
|
0.0447761194029851
|
0.0405405405405405
|
|
MIRNA
|
MIRNA:hsa-miR-199a-3p
|
hsa-miR-199a-3p
|
0.0395961397677493
|
114
|
88
|
4
|
0.0454545454545455
|
0.0350877192982456
|
|
MIRNA
|
MIRNA:hsa-miR-634
|
hsa-miR-634
|
0.039719662398578
|
71
|
71
|
3
|
0.0422535211267606
|
0.0422535211267606
|
|
MIRNA
|
MIRNA:hsa-miR-5585-5p
|
hsa-miR-5585-5p
|
0.041027411359911
|
62
|
83
|
3
|
0.036144578313253
|
0.0483870967741935
|
|
MIRNA
|
MIRNA:hsa-miR-186-5p
|
hsa-miR-186-5p
|
0.041027411359911
|
749
|
23
|
5
|
0.217391304347826
|
0.00667556742323097
|
|
MIRNA
|
MIRNA:hsa-miR-4777-5p
|
hsa-miR-4777-5p
|
0.041027411359911
|
55
|
94
|
3
|
0.0319148936170213
|
0.0545454545454545
|
|
MIRNA
|
MIRNA:hsa-miR-381-3p
|
hsa-miR-381-3p
|
0.041027411359911
|
135
|
76
|
4
|
0.0526315789473684
|
0.0296296296296296
|
|
MIRNA
|
MIRNA:hsa-miR-1179
|
hsa-miR-1179
|
0.0413579053720784
|
59
|
88
|
3
|
0.0340909090909091
|
0.0508474576271186
|
|
MIRNA
|
MIRNA:hsa-miR-4458
|
hsa-miR-4458
|
0.0416115923886602
|
310
|
54
|
5
|
0.0925925925925926
|
0.0161290322580645
|
|
MIRNA
|
MIRNA:hsa-miR-34a-5p
|
hsa-miR-34a-5p
|
0.0416115923886602
|
728
|
44
|
7
|
0.159090909090909
|
0.00961538461538462
|
|
MIRNA
|
MIRNA:hsa-miR-4500
|
hsa-miR-4500
|
0.0416115923886602
|
310
|
54
|
5
|
0.0925925925925926
|
0.0161290322580645
|
|
MIRNA
|
MIRNA:hsa-miR-4795-3p
|
hsa-miR-4795-3p
|
0.0431331891029622
|
122
|
86
|
4
|
0.0465116279069767
|
0.0327868852459016
|
|
MIRNA
|
MIRNA:hsa-miR-6865-3p
|
hsa-miR-6865-3p
|
0.0431331891029622
|
105
|
100
|
4
|
0.04
|
0.0380952380952381
|
|
MIRNA
|
MIRNA:hsa-miR-6825-5p
|
hsa-miR-6825-5p
|
0.0445468916255628
|
380
|
64
|
6
|
0.09375
|
0.0157894736842105
|
|
MIRNA
|
MIRNA:hsa-miR-429
|
hsa-miR-429
|
0.0447875470464958
|
151
|
71
|
4
|
0.0563380281690141
|
0.0264900662251656
|
|
MIRNA
|
MIRNA:hsa-miR-3663-3p
|
hsa-miR-3663-3p
|
0.046631101048046
|
56
|
100
|
3
|
0.03
|
0.0535714285714286
|
|
MIRNA
|
MIRNA:hsa-miR-7113-5p
|
hsa-miR-7113-5p
|
0.046631101048046
|
148
|
38
|
3
|
0.0789473684210526
|
0.0202702702702703
|
|
MIRNA
|
MIRNA:hsa-miR-6838-5p
|
hsa-miR-6838-5p
|
0.0466412693440559
|
431
|
95
|
8
|
0.0842105263157895
|
0.0185614849187935
|
|
MIRNA
|
MIRNA:hsa-miR-520c-5p
|
hsa-miR-520c-5p
|
0.047215596187552
|
61
|
94
|
3
|
0.0319148936170213
|
0.0491803278688525
|
|
MIRNA
|
MIRNA:hsa-miR-518d-5p
|
hsa-miR-518d-5p
|
0.047215596187552
|
61
|
94
|
3
|
0.0319148936170213
|
0.0491803278688525
|
|
MIRNA
|
MIRNA:hsa-miR-519c-5p
|
hsa-miR-519c-5p
|
0.047215596187552
|
61
|
94
|
3
|
0.0319148936170213
|
0.0491803278688525
|
|
MIRNA
|
MIRNA:hsa-miR-519b-5p
|
hsa-miR-519b-5p
|
0.047215596187552
|
61
|
94
|
3
|
0.0319148936170213
|
0.0491803278688525
|
|
MIRNA
|
MIRNA:hsa-miR-519a-5p
|
hsa-miR-519a-5p
|
0.047215596187552
|
61
|
94
|
3
|
0.0319148936170213
|
0.0491803278688525
|
|
MIRNA
|
MIRNA:hsa-miR-518e-5p
|
hsa-miR-518e-5p
|
0.047215596187552
|
61
|
94
|
3
|
0.0319148936170213
|
0.0491803278688525
|
|
MIRNA
|
MIRNA:hsa-miR-526a
|
hsa-miR-526a
|
0.047215596187552
|
61
|
94
|
3
|
0.0319148936170213
|
0.0491803278688525
|
|
MIRNA
|
MIRNA:hsa-miR-581
|
hsa-miR-581
|
0.047215596187552
|
57
|
100
|
3
|
0.03
|
0.0526315789473684
|
|
MIRNA
|
MIRNA:hsa-miR-518f-5p
|
hsa-miR-518f-5p
|
0.047215596187552
|
61
|
94
|
3
|
0.0319148936170213
|
0.0491803278688525
|
|
MIRNA
|
MIRNA:hsa-miR-523-5p
|
hsa-miR-523-5p
|
0.047215596187552
|
61
|
94
|
3
|
0.0319148936170213
|
0.0491803278688525
|
|
MIRNA
|
MIRNA:hsa-miR-4756-5p
|
hsa-miR-4756-5p
|
0.0475310060983421
|
284
|
40
|
4
|
0.1
|
0.0140845070422535
|
|
MIRNA
|
MIRNA:hsa-miR-224-5p
|
hsa-miR-224-5p
|
0.0475310060983421
|
132
|
86
|
4
|
0.0465116279069767
|
0.0303030303030303
|
|
MIRNA
|
MIRNA:hsa-miR-1321
|
hsa-miR-1321
|
0.0475310060983421
|
285
|
40
|
4
|
0.1
|
0.0140350877192982
|
|
MIRNA
|
MIRNA:hsa-miR-185-3p
|
hsa-miR-185-3p
|
0.0475310060983421
|
108
|
104
|
4
|
0.0384615384615385
|
0.037037037037037
|
|
MIRNA
|
MIRNA:hsa-miR-208a-5p
|
hsa-miR-208a-5p
|
0.0475310060983421
|
109
|
104
|
4
|
0.0384615384615385
|
0.036697247706422
|
|
MIRNA
|
MIRNA:hsa-let-7i-5p
|
hsa-let-7i-5p
|
0.0475310060983421
|
332
|
54
|
5
|
0.0925925925925926
|
0.0150602409638554
|
|
MIRNA
|
MIRNA:hsa-miR-921
|
hsa-miR-921
|
0.0475310060983421
|
68
|
85
|
3
|
0.0352941176470588
|
0.0441176470588235
|
|
MIRNA
|
MIRNA:hsa-miR-522-5p
|
hsa-miR-522-5p
|
0.0475310060983421
|
62
|
94
|
3
|
0.0319148936170213
|
0.0483870967741935
|
|
MIRNA
|
MIRNA:hsa-miR-4739
|
hsa-miR-4739
|
0.0478282158900249
|
288
|
40
|
4
|
0.1
|
0.0138888888888889
|
|
MIRNA
|
MIRNA:hsa-miR-208b-5p
|
hsa-miR-208b-5p
|
0.0480124768673687
|
110
|
104
|
4
|
0.0384615384615385
|
0.0363636363636364
|
|
MIRNA
|
MIRNA:hsa-miR-582-5p
|
hsa-miR-582-5p
|
0.0480124768673687
|
149
|
40
|
3
|
0.075
|
0.0201342281879195
|
|
MIRNA
|
MIRNA:hsa-miR-6715a-3p
|
hsa-miR-6715a-3p
|
0.0480124768673687
|
80
|
74
|
3
|
0.0405405405405405
|
0.0375
|
|
MIRNA
|
MIRNA:hsa-miR-92a-2-5p
|
hsa-miR-92a-2-5p
|
0.048239371151043
|
245
|
104
|
6
|
0.0576923076923077
|
0.0244897959183673
|
|
MIRNA
|
MIRNA:hsa-let-7g-5p
|
hsa-let-7g-5p
|
0.0484425759813344
|
339
|
54
|
5
|
0.0925925925925926
|
0.0147492625368732
|
|
MIRNA
|
MIRNA:hsa-miR-527
|
hsa-miR-527
|
0.0496874095794892
|
155
|
118
|
5
|
0.0423728813559322
|
0.032258064516129
|
|
MIRNA
|
MIRNA:hsa-miR-548ah-3p
|
hsa-miR-548ah-3p
|
0.0498612194996437
|
289
|
64
|
5
|
0.078125
|
0.0173010380622837
|
|
MIRNA
|
MIRNA:hsa-miR-548am-3p
|
hsa-miR-548am-3p
|
0.0498612194996437
|
289
|
64
|
5
|
0.078125
|
0.0173010380622837
|
|
MIRNA
|
MIRNA:hsa-miR-4645-3p
|
hsa-miR-4645-3p
|
0.0498612194996437
|
71
|
86
|
3
|
0.0348837209302326
|
0.0422535211267606
|
|
MIRNA
|
MIRNA:hsa-miR-548x-3p
|
hsa-miR-548x-3p
|
0.0499230252382428
|
291
|
64
|
5
|
0.078125
|
0.0171821305841924
|
|
MIRNA
|
MIRNA:hsa-miR-548ae-3p
|
hsa-miR-548ae-3p
|
0.0499230252382428
|
291
|
64
|
5
|
0.078125
|
0.0171821305841924
|
|
MIRNA
|
MIRNA:hsa-miR-548ag
|
hsa-miR-548ag
|
0.0499230252382428
|
73
|
84
|
3
|
0.0357142857142857
|
0.0410958904109589
|
|
MIRNA
|
MIRNA:hsa-miR-548aj-3p
|
hsa-miR-548aj-3p
|
0.0499230252382428
|
291
|
64
|
5
|
0.078125
|
0.0171821305841924
|
|
MIRNA
|
MIRNA:hsa-miR-548aq-3p
|
hsa-miR-548aq-3p
|
0.0499230252382428
|
291
|
64
|
5
|
0.078125
|
0.0171821305841924
|
|
MIRNA
|
MIRNA:hsa-miR-548j-3p
|
hsa-miR-548j-3p
|
0.0499230252382428
|
290
|
64
|
5
|
0.078125
|
0.0172413793103448
|
|
MIRNA
|
MIRNA:hsa-miR-498
|
hsa-miR-498
|
0.0499230252382428
|
316
|
59
|
5
|
0.0847457627118644
|
0.0158227848101266
|
|
REAC
|
REAC:R-HSA-9031628
|
NGF-stimulated transcription
|
8.53812706793888e-10
|
39
|
98
|
10
|
0.102040816326531
|
0.256410256410256
|
|
REAC
|
REAC:R-HSA-198725
|
Nuclear Events (kinase and transcription factor activation)
|
5.11963423382667e-08
|
61
|
98
|
10
|
0.102040816326531
|
0.163934426229508
|
|
REAC
|
REAC:R-HSA-187037
|
Signaling by NTRK1 (TRKA)
|
8.00752292199663e-06
|
112
|
20
|
6
|
0.3
|
0.0535714285714286
|
|
REAC
|
REAC:R-HSA-166520
|
Signaling by NTRKs
|
1.46639473979831e-05
|
130
|
20
|
6
|
0.3
|
0.0461538461538462
|
|
REAC
|
REAC:R-HSA-9018519
|
Estrogen-dependent gene expression
|
0.00181472812766632
|
112
|
6
|
3
|
0.5
|
0.0267857142857143
|
|
REAC
|
REAC:R-HSA-9006934
|
Signaling by Receptor Tyrosine Kinases
|
0.00181472812766632
|
485
|
5
|
4
|
0.8
|
0.00824742268041237
|
|
REAC
|
REAC:R-HSA-1257604
|
PIP3 activates AKT signaling
|
0.0034237125924213
|
249
|
29
|
6
|
0.206896551724138
|
0.0240963855421687
|
|
REAC
|
REAC:R-HSA-8939211
|
ESR-mediated signaling
|
0.00351064413273434
|
181
|
25
|
5
|
0.2
|
0.0276243093922652
|
|
REAC
|
REAC:R-HSA-937061
|
TRIF(TICAM1)-mediated TLR4 signaling
|
0.00563535736270865
|
96
|
35
|
4
|
0.114285714285714
|
0.0416666666666667
|
|
REAC
|
REAC:R-HSA-168176
|
Toll Like Receptor 5 (TLR5) Cascade
|
0.00563535736270865
|
84
|
35
|
4
|
0.114285714285714
|
0.0476190476190476
|
|
REAC
|
REAC:R-HSA-168164
|
Toll Like Receptor 3 (TLR3) Cascade
|
0.00563535736270865
|
92
|
35
|
4
|
0.114285714285714
|
0.0434782608695652
|
|
REAC
|
REAC:R-HSA-168181
|
Toll Like Receptor 7/8 (TLR7/8) Cascade
|
0.00563535736270865
|
91
|
35
|
4
|
0.114285714285714
|
0.043956043956044
|
|
REAC
|
REAC:R-HSA-162582
|
Signal Transduction
|
0.00563535736270865
|
2705
|
37
|
20
|
0.540540540540541
|
0.00739371534195933
|
|
REAC
|
REAC:R-HSA-181438
|
Toll Like Receptor 2 (TLR2) Cascade
|
0.00563535736270865
|
97
|
35
|
4
|
0.114285714285714
|
0.0412371134020619
|
|
REAC
|
REAC:R-HSA-168179
|
Toll Like Receptor TLR1:TLR2 Cascade
|
0.00563535736270865
|
97
|
35
|
4
|
0.114285714285714
|
0.0412371134020619
|
|
REAC
|
REAC:R-HSA-975138
|
TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation
|
0.00563535736270865
|
90
|
35
|
4
|
0.114285714285714
|
0.0444444444444444
|
|
REAC
|
REAC:R-HSA-166166
|
MyD88-independent TLR4 cascade
|
0.00563535736270865
|
96
|
35
|
4
|
0.114285714285714
|
0.0416666666666667
|
|
REAC
|
REAC:R-HSA-9006925
|
Intracellular signaling by second messengers
|
0.00563535736270865
|
289
|
29
|
6
|
0.206896551724138
|
0.0207612456747405
|
|
REAC
|
REAC:R-HSA-168142
|
Toll Like Receptor 10 (TLR10) Cascade
|
0.00563535736270865
|
84
|
35
|
4
|
0.114285714285714
|
0.0476190476190476
|
|
REAC
|
REAC:R-HSA-975155
|
MyD88 dependent cascade initiated on endosome
|
0.00563535736270865
|
91
|
35
|
4
|
0.114285714285714
|
0.043956043956044
|
|
REAC
|
REAC:R-HSA-166058
|
MyD88:MAL(TIRAP) cascade initiated on plasma membrane
|
0.00563535736270865
|
94
|
35
|
4
|
0.114285714285714
|
0.0425531914893617
|
|
REAC
|
REAC:R-HSA-168188
|
Toll Like Receptor TLR6:TLR2 Cascade
|
0.00563535736270865
|
94
|
35
|
4
|
0.114285714285714
|
0.0425531914893617
|
|
REAC
|
REAC:R-HSA-168138
|
Toll Like Receptor 9 (TLR9) Cascade
|
0.00563535736270865
|
95
|
35
|
4
|
0.114285714285714
|
0.0421052631578947
|
|
REAC
|
REAC:R-HSA-9006931
|
Signaling by Nuclear Receptors
|
0.00563535736270865
|
255
|
6
|
3
|
0.5
|
0.0117647058823529
|
|
REAC
|
REAC:R-HSA-975871
|
MyD88 cascade initiated on plasma membrane
|
0.00563535736270865
|
84
|
35
|
4
|
0.114285714285714
|
0.0476190476190476
|
|
REAC
|
REAC:R-HSA-1280215
|
Cytokine Signaling in Immune system
|
0.00591848687110562
|
842
|
9
|
5
|
0.555555555555556
|
0.00593824228028504
|
|
REAC
|
REAC:R-HSA-2262752
|
Cellular responses to stress
|
0.00906069294471358
|
546
|
29
|
7
|
0.241379310344828
|
0.0128205128205128
|
|
REAC
|
REAC:R-HSA-8953897
|
Cellular responses to external stimuli
|
0.0101694954971903
|
560
|
29
|
7
|
0.241379310344828
|
0.0125
|
|
REAC
|
REAC:R-HSA-6785807
|
Interleukin-4 and Interleukin-13 signaling
|
0.0101694954971903
|
110
|
17
|
3
|
0.176470588235294
|
0.0272727272727273
|
|
REAC
|
REAC:R-HSA-166016
|
Toll Like Receptor 4 (TLR4) Cascade
|
0.0109176821352226
|
126
|
35
|
4
|
0.114285714285714
|
0.0317460317460317
|
|
REAC
|
REAC:R-HSA-170834
|
Signaling by TGF-beta Receptor Complex
|
0.012508758771809
|
71
|
29
|
3
|
0.103448275862069
|
0.0422535211267606
|
|
REAC
|
REAC:R-HSA-3371497
|
HSP90 chaperone cycle for steroid hormone receptors (SHR)
|
0.0142140598902918
|
57
|
85
|
4
|
0.0470588235294118
|
0.0701754385964912
|
|
REAC
|
REAC:R-HSA-5675221
|
Negative regulation of MAPK pathway
|
0.0156173192758052
|
42
|
55
|
3
|
0.0545454545454545
|
0.0714285714285714
|
|
REAC
|
REAC:R-HSA-5683057
|
MAPK family signaling cascades
|
0.0156173192758052
|
308
|
29
|
5
|
0.172413793103448
|
0.0162337662337662
|
|
REAC
|
REAC:R-HSA-449147
|
Signaling by Interleukins
|
0.0156173192758052
|
446
|
6
|
3
|
0.5
|
0.00672645739910314
|
|
REAC
|
REAC:R-HSA-168898
|
Toll-like Receptor Cascades
|
0.0167726310988962
|
152
|
35
|
4
|
0.114285714285714
|
0.0263157894736842
|
|
REAC
|
REAC:R-HSA-450531
|
Regulation of mRNA stability by proteins that bind AU-rich elements
|
0.0175968131186636
|
86
|
29
|
3
|
0.103448275862069
|
0.0348837209302326
|
|
REAC
|
REAC:R-HSA-5687128
|
MAPK6/MAPK4 signaling
|
0.0186997841800036
|
89
|
29
|
3
|
0.103448275862069
|
0.0337078651685393
|
|
REAC
|
REAC:R-HSA-157118
|
Signaling by NOTCH
|
0.0238804637649378
|
197
|
31
|
4
|
0.129032258064516
|
0.0203045685279188
|
|
REAC
|
REAC:R-HSA-9006936
|
Signaling by TGFB family members
|
0.0238804637649378
|
100
|
29
|
3
|
0.103448275862069
|
0.03
|
|
REAC
|
REAC:R-HSA-2454202
|
Fc epsilon receptor (FCERI) signaling
|
0.0267873848848885
|
182
|
35
|
4
|
0.114285714285714
|
0.021978021978022
|
|
REAC
|
REAC:R-HSA-909733
|
Interferon alpha/beta signaling
|
0.0267873848848885
|
66
|
96
|
4
|
0.0416666666666667
|
0.0606060606060606
|
|
REAC
|
REAC:R-HSA-8856825
|
Cargo recognition for clathrin-mediated endocytosis
|
0.0307763601351698
|
104
|
32
|
3
|
0.09375
|
0.0288461538461538
|
|
REAC
|
REAC:R-HSA-3700989
|
Transcriptional Regulation by TP53
|
0.031979555999653
|
360
|
10
|
3
|
0.3
|
0.00833333333333333
|
|
REAC
|
REAC:R-HSA-212436
|
Generic Transcription Pathway
|
0.0348588477652732
|
1180
|
11
|
5
|
0.454545454545455
|
0.00423728813559322
|
|
REAC
|
REAC:R-HSA-8878159
|
Transcriptional regulation by RUNX3
|
0.0352508939351631
|
94
|
38
|
3
|
0.0789473684210526
|
0.0319148936170213
|
|
REAC
|
REAC:R-HSA-9006927
|
Signaling by Non-Receptor Tyrosine Kinases
|
0.0402541673742619
|
51
|
75
|
3
|
0.04
|
0.0588235294117647
|
|
REAC
|
REAC:R-HSA-8848021
|
Signaling by PTK6
|
0.0402541673742619
|
51
|
75
|
3
|
0.04
|
0.0588235294117647
|
|
REAC
|
REAC:R-HSA-6791312
|
TP53 Regulates Transcription of Cell Cycle Genes
|
0.0402541673742619
|
49
|
78
|
3
|
0.0384615384615385
|
0.0612244897959184
|
|
REAC
|
REAC:R-HSA-73857
|
RNA Polymerase II Transcription
|
0.0448451413811439
|
1301
|
11
|
5
|
0.454545454545455
|
0.00384319754035357
|
|
TF
|
TF:M00803_1
|
Factor: E2F; motif: GGCGSG; match class: 1
|
6.5739111782929e-09
|
10230
|
90
|
78
|
0.866666666666667
|
0.00762463343108504
|
|
TF
|
TF:M09826
|
Factor: BTEB3; motif: CCNNSCCNSCCCCKCCCCC
|
6.5739111782929e-09
|
11467
|
90
|
82
|
0.911111111111111
|
0.00715095491410133
|
|
TF
|
TF:M10529
|
Factor: Sp1; motif: RGGGMGGRGSNGGGG
|
6.5739111782929e-09
|
6891
|
96
|
67
|
0.697916666666667
|
0.00972282687563489
|
|
TF
|
TF:M01104_1
|
Factor: MOVO-B; motif: GNGGGGG; match class: 1
|
6.5739111782929e-09
|
5626
|
96
|
60
|
0.625
|
0.0106647707074298
|
|
TF
|
TF:M09723
|
Factor: BTEB1; motif: GGGGGCGGGGCNGSGGGNGS
|
1.54824935535477e-08
|
10156
|
89
|
76
|
0.853932584269663
|
0.00748326112642773
|
|
TF
|
TF:M07354
|
Factor: Egr-1; motif: GCGGGGGCGG
|
1.54824935535477e-08
|
7801
|
90
|
67
|
0.744444444444444
|
0.00858864248173311
|
|
TF
|
TF:M09826_1
|
Factor: BTEB3; motif: CCNNSCCNSCCCCKCCCCC; match class: 1
|
1.54824935535477e-08
|
7508
|
96
|
69
|
0.71875
|
0.00919019712306873
|
|
TF
|
TF:M10112
|
Factor: Miz-1; motif: NNRGGWGGGGGAGGGGMRR
|
3.27512114466386e-08
|
8711
|
96
|
74
|
0.770833333333333
|
0.00849500631385604
|
|
TF
|
TF:M11531
|
Factor: E2F-2; motif: GCGCGCGCGYW
|
3.27512114466386e-08
|
13270
|
90
|
86
|
0.955555555555556
|
0.00648078372268274
|
|
TF
|
TF:M07395_1
|
Factor: Sp1; motif: NGGGGCGGGGN; match class: 1
|
3.9324994074667e-08
|
6334
|
96
|
62
|
0.645833333333333
|
0.0097884433217556
|
|
TF
|
TF:M11531_1
|
Factor: E2F-2; motif: GCGCGCGCGYW; match class: 1
|
6.20868017960212e-08
|
12351
|
90
|
83
|
0.922222222222222
|
0.00672010363533317
|
|
TF
|
TF:M00691_1
|
Factor: ATF-1; motif: CYYTGACGTCA; match class: 1
|
7.79663419971956e-08
|
854
|
98
|
22
|
0.224489795918367
|
0.0257611241217799
|
|
TF
|
TF:M00986_1
|
Factor: Churchill; motif: CGGGNN; match class: 1
|
1.04890592545729e-07
|
10357
|
86
|
73
|
0.848837209302326
|
0.00704837308100801
|
|
TF
|
TF:M11529_1
|
Factor: E2F-2; motif: GCGCGCGCNCS; match class: 1
|
1.04890592545729e-07
|
14473
|
92
|
90
|
0.978260869565217
|
0.00621847578249154
|
|
TF
|
TF:M02036_1
|
Factor: WT1; motif: CGCCCCCNCN; match class: 1
|
1.22173601878217e-07
|
5290
|
89
|
52
|
0.584269662921348
|
0.00982986767485822
|
|
TF
|
TF:M02089
|
Factor: E2F-3; motif: GGCGGGN
|
1.22173601878217e-07
|
13173
|
89
|
84
|
0.943820224719101
|
0.00637667957185151
|
|
TF
|
TF:M09834
|
Factor: ZNF148; motif: NNNNNNCCNNCCCCTCCCCCACCCN
|
1.22173601878217e-07
|
6953
|
88
|
60
|
0.681818181818182
|
0.00862936861786279
|
|
TF
|
TF:M02036
|
Factor: WT1; motif: CGCCCCCNCN
|
1.79521959953902e-07
|
9767
|
90
|
73
|
0.811111111111111
|
0.00747414764001229
|
|
TF
|
TF:M12351_1
|
Factor: TIEG1; motif: NCCCNSNCCCCGCCCCC; match class: 1
|
2.09104341942916e-07
|
8180
|
88
|
65
|
0.738636363636364
|
0.00794621026894866
|
|
TF
|
TF:M08878
|
Factor: EGR; motif: CGCCCCCGCNN
|
2.09104341942916e-07
|
7483
|
90
|
63
|
0.7
|
0.00841908325537886
|
|
TF
|
TF:M01104
|
Factor: MOVO-B; motif: GNGGGGG
|
2.17802924600706e-07
|
10261
|
89
|
74
|
0.831460674157303
|
0.00721177273170256
|
|
TF
|
TF:M09973
|
Factor: CPBP; motif: GNNRGGGHGGGGNNGGGRN
|
2.63598489921835e-07
|
10853
|
96
|
81
|
0.84375
|
0.00746337418225375
|
|
TF
|
TF:M01820
|
Factor: CREM; motif: TGACGTCASYN
|
3.04928483883252e-07
|
6303
|
84
|
54
|
0.642857142857143
|
0.00856734888148501
|
|
TF
|
TF:M09984
|
Factor: MAZ; motif: GGGGGAGGGGGNGRGRRRGNRG
|
4.55093250803539e-07
|
9544
|
99
|
77
|
0.777777777777778
|
0.00806789606035205
|
|
TF
|
TF:M00803
|
Factor: E2F; motif: GGCGSG
|
4.55093250803539e-07
|
13190
|
90
|
84
|
0.933333333333333
|
0.00636846095526914
|
|
TF
|
TF:M12227_1
|
Factor: ZIC4; motif: NNCCNCCCRYNGYGN; match class: 1
|
4.55093250803539e-07
|
5898
|
96
|
57
|
0.59375
|
0.00966429298067141
|
|
TF
|
TF:M00196
|
Factor: Sp1; motif: NGGGGGCGGGGYN
|
6.36105021136012e-07
|
10233
|
89
|
73
|
0.820224719101124
|
0.00713378285937653
|
|
TF
|
TF:M12351
|
Factor: TIEG1; motif: NCCCNSNCCCCGCCCCC
|
6.62853046695569e-07
|
12302
|
90
|
81
|
0.9
|
0.0065842952365469
|
|
TF
|
TF:M04953_1
|
Factor: Sp1; motif: GGNDGGRGGCGGGG; match class: 1
|
6.62853046695569e-07
|
4065
|
85
|
42
|
0.494117647058824
|
0.0103321033210332
|
|
TF
|
TF:M12158
|
Factor: KLF15; motif: NCCMCGCCCMCN
|
7.14766743056349e-07
|
11088
|
89
|
76
|
0.853932584269663
|
0.00685425685425685
|
|
TF
|
TF:M03893_1
|
Factor: WT1; motif: GNGGGGGCGGGG; match class: 1
|
7.14766743056349e-07
|
3980
|
96
|
45
|
0.46875
|
0.0113065326633166
|
|
TF
|
TF:M10438_1
|
Factor: ZF5; motif: GGSGCGCGS; match class: 1
|
7.78897485035091e-07
|
14873
|
90
|
88
|
0.977777777777778
|
0.00591676191756875
|
|
TF
|
TF:M05327
|
Factor: WT1; motif: NGCGGGGGGGTSMMCYN
|
7.78897485035091e-07
|
5509
|
98
|
55
|
0.561224489795918
|
0.00998366309675077
|
|
TF
|
TF:M11022_1
|
Factor: IRX2a; motif: ACRYGNNNNACRYGT; match class: 1
|
8.34398760514334e-07
|
6246
|
89
|
55
|
0.617977528089888
|
0.00880563560678834
|
|
TF
|
TF:M00932
|
Factor: Sp1; motif: NNGGGGCGGGGNN
|
9.6909854755378e-07
|
10419
|
96
|
78
|
0.8125
|
0.00748632306363375
|
|
TF
|
TF:M07395
|
Factor: Sp1; motif: NGGGGCGGGGN
|
1.01039341525914e-06
|
10640
|
89
|
74
|
0.831460674157303
|
0.00695488721804511
|
|
TF
|
TF:M02089_1
|
Factor: E2F-3; motif: GGCGGGN; match class: 1
|
1.01039341525914e-06
|
9362
|
89
|
69
|
0.775280898876405
|
0.00737022003845332
|
|
TF
|
TF:M09761
|
Factor: EGR1; motif: NGNGKGGGYGGNGS
|
1.10399685500818e-06
|
5162
|
89
|
49
|
0.550561797752809
|
0.00949244478884153
|
|
TF
|
TF:M01857
|
Factor: AP-2alpha; motif: NGCCYSNNGSN
|
1.12872785934895e-06
|
9178
|
84
|
65
|
0.773809523809524
|
0.00708215297450425
|
|
TF
|
TF:M01873
|
Factor: Egr-1; motif: GCGGGGGCGG
|
1.21012476330762e-06
|
6738
|
89
|
57
|
0.640449438202247
|
0.00845948352626892
|
|
TF
|
TF:M12160
|
Factor: KLF15; motif: RCCMCRCCCMCN
|
1.21012476330762e-06
|
12729
|
89
|
81
|
0.910112359550562
|
0.00636342210699976
|
|
TF
|
TF:M03876_1
|
Factor: Kaiso; motif: GCMGGGRGCRGS; match class: 1
|
1.35238945196787e-06
|
9106
|
88
|
67
|
0.761363636363636
|
0.00735778607511531
|
|
TF
|
TF:M03893
|
Factor: WT1; motif: GNGGGGGCGGGG
|
1.4578622365464e-06
|
8290
|
96
|
68
|
0.708333333333333
|
0.00820265379975875
|
|
TF
|
TF:M00933
|
Factor: Sp1; motif: CCCCGCCCCN
|
1.50091203586334e-06
|
9715
|
89
|
70
|
0.786516853932584
|
0.00720535254760679
|
|
TF
|
TF:M01303
|
Factor: SP1; motif: GGGGYGGGGNS
|
1.5691272056598e-06
|
7898
|
96
|
66
|
0.6875
|
0.00835654596100279
|
|
TF
|
TF:M07040_1
|
Factor: GKLF; motif: NNRRGRRNGNSNNN; match class: 1
|
1.59353901764161e-06
|
8156
|
101
|
70
|
0.693069306930693
|
0.00858263854830799
|
|
TF
|
TF:M07397_1
|
Factor: ZBP89; motif: CCCCKCCCCCNN; match class: 1
|
1.6808325129564e-06
|
2951
|
88
|
35
|
0.397727272727273
|
0.0118603863097255
|
|
TF
|
TF:M08878_1
|
Factor: EGR; motif: CGCCCCCGCNN; match class: 1
|
1.74762661647842e-06
|
3224
|
89
|
37
|
0.415730337078652
|
0.0114764267990074
|
|
TF
|
TF:M09894_1
|
Factor: E2F-4; motif: SNGGGCGGGAANN; match class: 1
|
1.93163826545336e-06
|
13929
|
90
|
85
|
0.944444444444444
|
0.00610237633713834
|
|
TF
|
TF:M04869_1
|
Factor: Egr-1; motif: GCGCATGCG; match class: 1
|
2.40407466437456e-06
|
10253
|
85
|
69
|
0.811764705882353
|
0.00672973763776456
|
|
TF
|
TF:M07250_1
|
Factor: E2F-1; motif: NNNSSCGCSAANN; match class: 1
|
2.50852150514966e-06
|
5515
|
89
|
50
|
0.561797752808989
|
0.00906618313689937
|
|
TF
|
TF:M00716_1
|
Factor: ZF5; motif: GSGCGCGR; match class: 1
|
2.61844990205572e-06
|
14106
|
92
|
87
|
0.945652173913043
|
0.00616758826031476
|
|
TF
|
TF:M09834_1
|
Factor: ZNF148; motif: NNNNNNCCNNCCCCTCCCCCACCCN; match class: 1
|
2.70810805773559e-06
|
2869
|
96
|
36
|
0.375
|
0.0125479261066574
|
|
TF
|
TF:M12313
|
Factor: ZNF460; motif: NNACNCCCCCCNN
|
3.01188907332323e-06
|
5825
|
88
|
51
|
0.579545454545455
|
0.00875536480686695
|
|
TF
|
TF:M11882_1
|
Factor: pax-6; motif: NYACGCNYSANYGMNCN; match class: 1
|
3.49578348065478e-06
|
11423
|
88
|
75
|
0.852272727272727
|
0.00656570077912983
|
|
TF
|
TF:M04869
|
Factor: Egr-1; motif: GCGCATGCG
|
3.53037145772956e-06
|
11232
|
96
|
80
|
0.833333333333333
|
0.00712250712250712
|
|
TF
|
TF:M02281_1
|
Factor: SP1; motif: CCCCKCCCCC; match class: 1
|
3.5539011618183e-06
|
2558
|
126
|
40
|
0.317460317460317
|
0.0156372165754496
|
|
TF
|
TF:M09887_1
|
Factor: CREB1; motif: NRRTGACGTMA; match class: 1
|
3.6850450861087e-06
|
1616
|
72
|
22
|
0.305555555555556
|
0.0136138613861386
|
|
TF
|
TF:M01837
|
Factor: FKLF; motif: BGGGNGGVMD
|
3.6850450861087e-06
|
5442
|
96
|
52
|
0.541666666666667
|
0.0095553105475928
|
|
TF
|
TF:M03989
|
Factor: FLI1; motif: ACCGGAAATCCGGT
|
3.6850450861087e-06
|
10442
|
95
|
76
|
0.8
|
0.00727829917640299
|
|
TF
|
TF:M02281
|
Factor: SP1; motif: CCCCKCCCCC
|
3.79434875745692e-06
|
6480
|
88
|
54
|
0.613636363636364
|
0.00833333333333333
|
|
TF
|
TF:M08911
|
Factor: CTCF; motif: NCCRSTAGGGGGCGC
|
4.09686688152683e-06
|
9094
|
90
|
67
|
0.744444444444444
|
0.00736749505168243
|
|
TF
|
TF:M09970
|
Factor: KLF3; motif: NNNNNNGGGCGGGGCNNGN
|
4.14036720858097e-06
|
7728
|
96
|
64
|
0.666666666666667
|
0.0082815734989648
|
|
TF
|
TF:M00113
|
Factor: CREB; motif: NNGNTGACGTNN
|
4.14036720858097e-06
|
4801
|
85
|
44
|
0.517647058823529
|
0.00916475734222037
|
|
TF
|
TF:M07312
|
Factor: ATF-2; motif: NNTGACGTCAN
|
4.14036720858097e-06
|
3483
|
83
|
36
|
0.433734939759036
|
0.0103359173126615
|
|
TF
|
TF:M10072
|
Factor: sp4; motif: NNGNARGRGGCGGRGCNNRR
|
4.14036720858097e-06
|
10494
|
89
|
72
|
0.808988764044944
|
0.00686106346483705
|
|
TF
|
TF:M04148
|
Factor: TFAP2A; motif: NGCCCYNNGGGCN
|
4.14036720858097e-06
|
10040
|
101
|
78
|
0.772277227722772
|
0.00776892430278884
|
|
TF
|
TF:M04148_1
|
Factor: TFAP2A; motif: NGCCCYNNGGGCN; match class: 1
|
4.14383733442621e-06
|
9613
|
101
|
76
|
0.752475247524752
|
0.00790596067824821
|
|
TF
|
TF:M07206_1
|
Factor: E2F-1; motif: NGGGCGGGARV; match class: 1
|
4.18871984241306e-06
|
10849
|
90
|
74
|
0.822222222222222
|
0.00682090515254862
|
|
TF
|
TF:M07397
|
Factor: ZBP89; motif: CCCCKCCCCCNN
|
4.18871984241306e-06
|
7143
|
88
|
57
|
0.647727272727273
|
0.00797984040319194
|
|
TF
|
TF:M07354_1
|
Factor: Egr-1; motif: GCGGGGGCGG; match class: 1
|
5.009930937729e-06
|
3561
|
89
|
38
|
0.426966292134831
|
0.0106711597865768
|
|
TF
|
TF:M00981
|
Factor: CREB,; motif: NTGACGTNA
|
5.17528730502578e-06
|
8345
|
84
|
60
|
0.714285714285714
|
0.00718993409227082
|
|
TF
|
TF:M10432
|
Factor: MAZ; motif: GGGMGGGGS
|
5.40236645687154e-06
|
9259
|
96
|
71
|
0.739583333333333
|
0.00766821471001188
|
|
TF
|
TF:M03807
|
Factor: SP2; motif: GNNGGGGGCGGGGSN
|
5.65395281511398e-06
|
8415
|
96
|
67
|
0.697916666666667
|
0.00796197266785502
|
|
TF
|
TF:M07248
|
Factor: CREB1; motif: NNNNTGACGTNANNN
|
6.10455876724724e-06
|
4266
|
83
|
40
|
0.481927710843373
|
0.00937646507266761
|
|
TF
|
TF:M12160_1
|
Factor: KLF15; motif: RCCMCRCCCMCN; match class: 1
|
6.43959109861797e-06
|
8031
|
96
|
65
|
0.677083333333333
|
0.00809363715602042
|
|
TF
|
TF:M01873_1
|
Factor: Egr-1; motif: GCGGGGGCGG; match class: 1
|
6.45264577793422e-06
|
2672
|
89
|
32
|
0.359550561797753
|
0.0119760479041916
|
|
TF
|
TF:M03544
|
Factor: CREB1; motif: NNNNACGTCANN
|
6.61756081952041e-06
|
4101
|
83
|
39
|
0.469879518072289
|
0.00950987564008778
|
|
TF
|
TF:M10112_1
|
Factor: Miz-1; motif: NNRGGWGGGGGAGGGGMRR; match class: 1
|
6.73393642170293e-06
|
4075
|
96
|
43
|
0.447916666666667
|
0.0105521472392638
|
|
TF
|
TF:M06444
|
Factor: ZNF557; motif: NCCGCKTCCTGC
|
6.77170555953888e-06
|
1344
|
89
|
22
|
0.247191011235955
|
0.0163690476190476
|
|
TF
|
TF:M01783
|
Factor: SP2; motif: GGGCGGGAC
|
6.77170555953888e-06
|
9148
|
89
|
66
|
0.741573033707865
|
0.00721469173589856
|
|
TF
|
TF:M11882
|
Factor: pax-6; motif: NYACGCNYSANYGMNCN
|
8.49514092704881e-06
|
15490
|
90
|
88
|
0.977777777777778
|
0.00568108457069077
|
|
TF
|
TF:M07208
|
Factor: EGR1; motif: NCNCCGCCCCCGCN
|
8.69079517432605e-06
|
6183
|
89
|
52
|
0.584269662921348
|
0.0084101568817726
|
|
TF
|
TF:M03567
|
Factor: Sp2; motif: NYSGCCCCGCCCCCY
|
9.22406904786077e-06
|
7710
|
96
|
63
|
0.65625
|
0.00817120622568093
|
|
TF
|
TF:M09894
|
Factor: E2F-4; motif: SNGGGCGGGAANN
|
9.26151051801487e-06
|
16194
|
96
|
95
|
0.989583333333333
|
0.00586637026059034
|
|
TF
|
TF:M12158_1
|
Factor: KLF15; motif: NCCMCGCCCMCN; match class: 1
|
9.26768770752079e-06
|
6004
|
89
|
51
|
0.573033707865168
|
0.00849433710859427
|
|
TF
|
TF:M03895_1
|
Factor: CTCF; motif: WGCGCCMYCTAGYGGYN; match class: 1
|
9.30362807114497e-06
|
3511
|
96
|
39
|
0.40625
|
0.0111079464540017
|
|
TF
|
TF:M07380
|
Factor: E2F-4; motif: NTTTCSCGCC
|
9.32926012648024e-06
|
13022
|
85
|
77
|
0.905882352941176
|
0.00591307018891107
|
|
TF
|
TF:M06948
|
Factor: Sp2; motif: TGGGCGCGCCCA
|
9.57976971150194e-06
|
8864
|
90
|
65
|
0.722222222222222
|
0.00733303249097473
|
|
TF
|
TF:M00178
|
Factor: CREB; motif: NSTGACGTMANN
|
1.00739951570365e-05
|
5255
|
84
|
45
|
0.535714285714286
|
0.00856327307326356
|
|
TF
|
TF:M00716
|
Factor: ZF5; motif: GSGCGCGR
|
1.12361971739925e-05
|
16103
|
92
|
91
|
0.989130434782609
|
0.00565112090914736
|
|
TF
|
TF:M00933_1
|
Factor: Sp1; motif: CCCCGCCCCN; match class: 1
|
1.14874224917758e-05
|
5166
|
96
|
49
|
0.510416666666667
|
0.00948509485094851
|
|
TF
|
TF:M00695
|
Factor: ETF; motif: GVGGMGG
|
1.18635308268785e-05
|
10430
|
88
|
70
|
0.795454545454545
|
0.00671140939597315
|
|
TF
|
TF:M10026
|
Factor: PATZ; motif: GGGGNGGGGGMKGGRRNGGNRN
|
1.1952750204466e-05
|
8389
|
96
|
66
|
0.6875
|
0.00786744546429849
|
|
TF
|
TF:M01240_1
|
Factor: BEN; motif: CAGCGRNV; match class: 1
|
1.20127905602076e-05
|
13227
|
92
|
83
|
0.902173913043478
|
0.0062750434716867
|
|
TF
|
TF:M00982
|
Factor: KROX; motif: CCCGCCCCCRCCCC
|
1.20127905602076e-05
|
7815
|
90
|
60
|
0.666666666666667
|
0.00767754318618042
|
|
TF
|
TF:M09866
|
Factor: ATF-1; motif: NNRTGACGYMA
|
1.20127905602076e-05
|
3162
|
72
|
30
|
0.416666666666667
|
0.0094876660341556
|
|
TF
|
TF:M11529
|
Factor: E2F-2; motif: GCGCGCGCNCS
|
1.20127905602076e-05
|
16263
|
96
|
95
|
0.989583333333333
|
0.00584148066162455
|
|
TF
|
TF:M12155
|
Factor: Sp2; motif: NTWAGTCCCGCCCMCTT
|
1.21376891187156e-05
|
4591
|
86
|
42
|
0.488372093023256
|
0.00914833369636245
|
|
TF
|
TF:M00931
|
Factor: Sp1; motif: GGGGCGGGGC
|
1.21376891187156e-05
|
10281
|
89
|
70
|
0.786516853932584
|
0.00680867619881334
|
|
TF
|
TF:M11603_1
|
Factor: TCF-1; motif: ACATCGRGRCGCTGW; match class: 1
|
1.23084827758736e-05
|
13184
|
96
|
86
|
0.895833333333333
|
0.00652305825242718
|
|
TF
|
TF:M09636_1
|
Factor: MAZ; motif: GGGMGGGGSSGGGGGGGGGGGG; match class: 1
|
1.31183353735574e-05
|
14096
|
90
|
84
|
0.933333333333333
|
0.00595913734392736
|
|
TF
|
TF:M00017
|
Factor: ATF; motif: CNSTGACGTNNNYC
|
1.3391663133553e-05
|
7226
|
98
|
61
|
0.622448979591837
|
0.00844173816772765
|
|
TF
|
TF:M00053
|
Factor: c-Rel; motif: SGGRNTTTCC
|
1.34809866404102e-05
|
3672
|
126
|
48
|
0.380952380952381
|
0.0130718954248366
|
|
TF
|
TF:M00470
|
Factor: AP-2gamma; motif: GCCYNNGGS
|
1.39284273635707e-05
|
9422
|
95
|
70
|
0.736842105263158
|
0.00742942050520059
|
|
TF
|
TF:M09592
|
Factor: ATF-1; motif: NRTGACGTMAN
|
1.42789418502009e-05
|
4411
|
72
|
36
|
0.5
|
0.00816141464520517
|
|
TF
|
TF:M00470_1
|
Factor: AP-2gamma; motif: GCCYNNGGS; match class: 1
|
1.61143442072296e-05
|
5600
|
86
|
47
|
0.546511627906977
|
0.00839285714285714
|
|
TF
|
TF:M04953
|
Factor: Sp1; motif: GGNDGGRGGCGGGG
|
1.67371214316267e-05
|
8688
|
96
|
67
|
0.697916666666667
|
0.00771178637200737
|
|
TF
|
TF:M08525_1
|
Factor: E2F-1:HES-7; motif: GGCRCGTGSYNNWNGGCGCSM; match class: 1
|
1.76682525656667e-05
|
15268
|
90
|
87
|
0.966666666666667
|
0.00569819229761593
|
|
TF
|
TF:M07226
|
Factor: SP1; motif: NCCCCKCCCCC
|
1.80881398857199e-05
|
8292
|
96
|
65
|
0.677083333333333
|
0.0078388808490111
|
|
TF
|
TF:M10438
|
Factor: ZF5; motif: GGSGCGCGS
|
1.80881398857199e-05
|
16228
|
92
|
91
|
0.989130434782609
|
0.00560759181661326
|
|
TF
|
TF:M11595
|
Factor: GCMa; motif: RTGCGGGTN
|
1.82465365955206e-05
|
12117
|
83
|
72
|
0.867469879518072
|
0.00594206486754147
|
|
TF
|
TF:M10072_1
|
Factor: sp4; motif: NNGNARGRGGCGGRGCNNRR; match class: 1
|
1.9759277080045e-05
|
6225
|
84
|
49
|
0.583333333333333
|
0.0078714859437751
|
|
TF
|
TF:M07039
|
Factor: ETF; motif: CCCCGCCCCYN
|
1.99142737626476e-05
|
16384
|
96
|
95
|
0.989583333333333
|
0.00579833984375
|
|
TF
|
TF:M04515
|
Factor: E2F-1; motif: WWTGGCGCCAAA
|
2.28957810512933e-05
|
13334
|
86
|
78
|
0.906976744186046
|
0.00584970751462427
|
|
TF
|
TF:M11883_1
|
Factor: pax-6; motif: NYACGCNTSRNYGCNYN; match class: 1
|
2.37354431235899e-05
|
7768
|
90
|
59
|
0.655555555555556
|
0.00759526261585994
|
|
TF
|
TF:M09658
|
Factor: Sp2; motif: GGSNNGGGGGCGGGGCCNGNGS
|
2.56511362102154e-05
|
5262
|
84
|
44
|
0.523809523809524
|
0.00836183960471304
|
|
TF
|
TF:M01820_1
|
Factor: CREM; motif: TGACGTCASYN; match class: 1
|
2.62591188200493e-05
|
1879
|
72
|
22
|
0.305555555555556
|
0.0117083555082491
|
|
TF
|
TF:M01118
|
Factor: WT1; motif: SMCNCCNSC
|
2.80450525065465e-05
|
6808
|
96
|
57
|
0.59375
|
0.00837250293772033
|
|
TF
|
TF:M00244
|
Factor: NGFI-C; motif: WTGCGTGGGYGG
|
2.80450525065465e-05
|
6184
|
83
|
48
|
0.578313253012048
|
0.00776196636481242
|
|
TF
|
TF:M00931_1
|
Factor: Sp1; motif: GGGGCGGGGC; match class: 1
|
3.12760601701145e-05
|
5905
|
96
|
52
|
0.541666666666667
|
0.00880609652836579
|
|
TF
|
TF:M12173
|
Factor: GKLF; motif: NNCCMCRCCCN
|
3.12760601701145e-05
|
10023
|
89
|
68
|
0.764044943820225
|
0.00678439588945426
|
|
TF
|
TF:M09896
|
Factor: E2F-7; motif: GRGGCGGGAANNN
|
3.19508379483582e-05
|
9927
|
96
|
72
|
0.75
|
0.00725294650951949
|
|
TF
|
TF:M09897
|
Factor: Egr-1; motif: NNNNGCGKGGGYGGNRN
|
3.29122984222062e-05
|
4244
|
89
|
40
|
0.449438202247191
|
0.00942507068803016
|
|
TF
|
TF:M00196_1
|
Factor: Sp1; motif: NGGGGGCGGGGYN; match class: 1
|
3.30286880955824e-05
|
5919
|
96
|
52
|
0.541666666666667
|
0.00878526778171988
|
|
TF
|
TF:M00144_1
|
Factor: Pax-5; motif: RRMSWGANWYCTNRAGCGKRACSRYNSM; match class: 1
|
3.60399959391507e-05
|
7128
|
89
|
55
|
0.617977528089888
|
0.00771604938271605
|
|
TF
|
TF:M11391
|
Factor: Erg; motif: NACCGGATATCCGGTN
|
3.61322535371765e-05
|
10082
|
92
|
70
|
0.760869565217391
|
0.00694306685181512
|
|
TF
|
TF:M01864
|
Factor: ATF-4; motif: KACGTCAKS
|
3.66500239928654e-05
|
4477
|
31
|
20
|
0.645161290322581
|
0.00446727719454992
|
|
TF
|
TF:M11601
|
Factor: TCF-1; motif: ACATCGRGRCGCTGW
|
3.67940339168942e-05
|
14823
|
85
|
81
|
0.952941176470588
|
0.00546448087431694
|
|
TF
|
TF:M08441_1
|
Factor: AP-2gamma:Elk-1; motif: NGCCKNRGGSGRCGGAAGTG; match class: 1
|
3.67940339168942e-05
|
12761
|
85
|
75
|
0.882352941176471
|
0.00587728234464384
|
|
TF
|
TF:M08487_1
|
Factor: GCMa:Erg; motif: ATGCGGGCGGAARKG; match class: 1
|
3.67940339168942e-05
|
8790
|
88
|
62
|
0.704545454545455
|
0.00705346985210466
|
|
TF
|
TF:M03896
|
Factor: EGR1; motif: NACGCCCACGCANW
|
3.69802743408322e-05
|
6129
|
89
|
50
|
0.561797752808989
|
0.00815793767335618
|
|
TF
|
TF:M07226_1
|
Factor: SP1; motif: NCCCCKCCCCC; match class: 1
|
3.76452074762968e-05
|
3915
|
96
|
40
|
0.416666666666667
|
0.0102171136653895
|
|
TF
|
TF:M10108
|
Factor: WT1; motif: RGGNGGGGGAGGRGGNGGRG
|
3.8828565907161e-05
|
6511
|
96
|
55
|
0.572916666666667
|
0.00844724312701582
|
|
TF
|
TF:M09898_1
|
Factor: Egr-2; motif: GNGRRNGWGKGGGNGGRG; match class: 1
|
4.12053645365174e-05
|
2541
|
88
|
29
|
0.329545454545455
|
0.0114128295946478
|
|
TF
|
TF:M07277
|
Factor: BTEB2; motif: RGGGNGKGGN
|
4.12606242763071e-05
|
8492
|
96
|
65
|
0.677083333333333
|
0.00765426283560999
|
|
TF
|
TF:M10071
|
Factor: Sp1; motif: NGGGGGCGGGGCCNGGGGGGGG
|
4.15813797038482e-05
|
8495
|
96
|
65
|
0.677083333333333
|
0.00765155974102413
|
|
TF
|
TF:M03895
|
Factor: CTCF; motif: WGCGCCMYCTAGYGGYN
|
4.1896022865464e-05
|
9251
|
90
|
65
|
0.722222222222222
|
0.00702626743054805
|
|
TF
|
TF:M04823_1
|
Factor: E2F-4; motif: NNTTCCCGCCNN; match class: 1
|
4.24750466139938e-05
|
7850
|
83
|
55
|
0.662650602409639
|
0.00700636942675159
|
|
TF
|
TF:M12140_1
|
Factor: Egr-1; motif: NNCRCCCMCGCNN; match class: 1
|
4.35026702451232e-05
|
1561
|
95
|
23
|
0.242105263157895
|
0.0147341447789878
|
|
TF
|
TF:M10432_1
|
Factor: MAZ; motif: GGGMGGGGS; match class: 1
|
4.43240479658528e-05
|
4365
|
82
|
38
|
0.463414634146341
|
0.00870561282932417
|
|
TF
|
TF:M08441
|
Factor: AP-2gamma:Elk-1; motif: NGCCKNRGGSGRCGGAAGTG
|
4.49158353488629e-05
|
16345
|
89
|
88
|
0.98876404494382
|
0.00538390945243194
|
|
TF
|
TF:M09892_1
|
Factor: E2F-1; motif: NNNNGGCGGGAARN; match class: 1
|
5.28895451520752e-05
|
9162
|
86
|
62
|
0.720930232558139
|
0.00676708142327003
|
|
TF
|
TF:M11058_1
|
Factor: Hey1; motif: NGCRCGYGYN; match class: 1
|
5.31809897552975e-05
|
10925
|
89
|
71
|
0.797752808988764
|
0.00649885583524027
|
|
TF
|
TF:M04107
|
Factor: RUNX2; motif: WRACCGCANWAACCGCAN
|
5.71275340501203e-05
|
12351
|
86
|
74
|
0.86046511627907
|
0.00599141769897174
|
|
TF
|
TF:M05432
|
Factor: ZSCAN18; motif: TRRCGRCGNMCC
|
5.71275340501203e-05
|
4117
|
87
|
38
|
0.436781609195402
|
0.00923002186057809
|
|
TF
|
TF:M00810_1
|
Factor: SRF; motif: SCCAWATAWGGMNMNNNN; match class: 1
|
5.72761522500234e-05
|
7
|
68
|
3
|
0.0441176470588235
|
0.428571428571429
|
|
TF
|
TF:M00932_1
|
Factor: Sp1; motif: NNGGGGCGGGGNN; match class: 1
|
5.74824055052754e-05
|
6047
|
96
|
52
|
0.541666666666667
|
0.0085993054407144
|
|
TF
|
TF:M11697_1
|
Factor: MEF-2B1; motif: CCNWATWNRG; match class: 1
|
5.93177467520975e-05
|
683
|
4
|
4
|
1
|
0.00585651537335286
|
|
TF
|
TF:M10426_1
|
Factor: ctcf; motif: CCRSCAGGGGGCGCN; match class: 1
|
5.97086672765594e-05
|
4369
|
89
|
40
|
0.449438202247191
|
0.00915541313801785
|
|
TF
|
TF:M00178_1
|
Factor: CREB; motif: NSTGACGTMANN; match class: 1
|
5.98834405052844e-05
|
1522
|
72
|
19
|
0.263888888888889
|
0.0124835742444152
|
|
TF
|
TF:M01199
|
Factor: RNF96; motif: BCCCGCRGCC
|
6.14490152397573e-05
|
8473
|
88
|
60
|
0.681818181818182
|
0.00708131712498525
|
|
TF
|
TF:M00981_1
|
Factor: CREB,; motif: NTGACGTNA; match class: 1
|
6.58139423588584e-05
|
2904
|
72
|
27
|
0.375
|
0.00929752066115703
|
|
TF
|
TF:M03896_1
|
Factor: EGR1; motif: NACGCCCACGCANW; match class: 1
|
6.58139423588584e-05
|
2106
|
85
|
25
|
0.294117647058824
|
0.0118708452041785
|
|
TF
|
TF:M04826_1
|
Factor: p300; motif: ACNTCCG; match class: 1
|
6.58139423588584e-05
|
15260
|
99
|
94
|
0.94949494949495
|
0.00615989515072084
|
|
TF
|
TF:M00916
|
Factor: CREB; motif: NNTKACGTCANNNS
|
6.58139423588584e-05
|
5889
|
84
|
46
|
0.547619047619048
|
0.00781117337408728
|
|
TF
|
TF:M00333_1
|
Factor: ZF5; motif: NRNGNGCGCGCWN; match class: 1
|
6.62192796513551e-05
|
12401
|
90
|
77
|
0.855555555555556
|
0.00620917667930006
|
|
TF
|
TF:M09984_1
|
Factor: MAZ; motif: GGGGGAGGGGGNGRGRRRGNRG; match class: 1
|
7.00600734528121e-05
|
5555
|
96
|
49
|
0.510416666666667
|
0.00882088208820882
|
|
TF
|
TF:M01118_1
|
Factor: WT1; motif: SMCNCCNSC; match class: 1
|
7.18035856020994e-05
|
2766
|
63
|
24
|
0.380952380952381
|
0.00867678958785249
|
|
TF
|
TF:M11393_1
|
Factor: Erg; motif: NACCGGATATCCGGTN; match class: 1
|
7.46570292308317e-05
|
13665
|
86
|
78
|
0.906976744186046
|
0.00570801317233809
|
|
TF
|
TF:M07084_1
|
Factor: E2F-4; motif: NGGCGGGAARN; match class: 1
|
7.54885491599662e-05
|
5437
|
85
|
44
|
0.517647058823529
|
0.00809269817914291
|
|
TF
|
TF:M09592_1
|
Factor: ATF-1; motif: NRTGACGTMAN; match class: 1
|
7.57297341162063e-05
|
1255
|
72
|
17
|
0.236111111111111
|
0.0135458167330677
|
|
TF
|
TF:M00800
|
Factor: AP-2; motif: GSCCSCRGGCNRNRNN
|
8.24365474511598e-05
|
9119
|
101
|
70
|
0.693069306930693
|
0.00767628029389187
|
|
TF
|
TF:M03876
|
Factor: Kaiso; motif: GCMGGGRGCRGS
|
8.24365474511598e-05
|
13622
|
90
|
81
|
0.9
|
0.00594626339744531
|
|
TF
|
TF:M07034
|
Factor: ATF-1; motif: NNNTGACGTNNN
|
8.24365474511598e-05
|
4248
|
83
|
37
|
0.44578313253012
|
0.00870998116760829
|
|
TF
|
TF:M00986
|
Factor: Churchill; motif: CGGGNN
|
8.24365474511598e-05
|
14197
|
89
|
82
|
0.921348314606742
|
0.00577586814115658
|
|
TF
|
TF:M04515_1
|
Factor: E2F-1; motif: WWTGGCGCCAAA; match class: 1
|
8.24365474511598e-05
|
12173
|
86
|
73
|
0.848837209302326
|
0.00599687833730387
|
|
TF
|
TF:M09765
|
Factor: SP1; motif: NRGKGGGCGGGGCN
|
8.44562391238106e-05
|
7469
|
96
|
59
|
0.614583333333333
|
0.00789931717766769
|
|
TF
|
TF:M03914
|
Factor: KLF16; motif: GMCACGCCCCC
|
8.46742885463575e-05
|
3363
|
98
|
36
|
0.36734693877551
|
0.0107047279214987
|
|
TF
|
TF:M11058
|
Factor: Hey1; motif: NGCRCGYGYN
|
9.64971682094355e-05
|
13162
|
96
|
84
|
0.875
|
0.00638200881325027
|
|
TF
|
TF:M07039_1
|
Factor: ETF; motif: CCCCGCCCCYN; match class: 1
|
9.90695020931675e-05
|
13611
|
89
|
80
|
0.898876404494382
|
0.00587759900080817
|
|
TF
|
TF:M11883
|
Factor: pax-6; motif: NYACGCNTSRNYGCNYN
|
0.000100253745888421
|
13445
|
96
|
85
|
0.885416666666667
|
0.00632205280773522
|
|
TF
|
TF:M01861
|
Factor: ATF-1; motif: TNACGTCAN
|
0.000100679967110946
|
6294
|
83
|
47
|
0.566265060240964
|
0.00746742929774388
|
|
TF
|
TF:M00152
|
Factor: SRF; motif: ATGCCCATATATGGWNNT
|
0.00010876076501488
|
182
|
4
|
3
|
0.75
|
0.0164835164835165
|
|
TF
|
TF:M04106
|
Factor: RUNX2; motif: NRACCGCAAACCGCAN
|
0.000109541610536756
|
13833
|
87
|
79
|
0.908045977011494
|
0.00571098098749367
|
|
TF
|
TF:M08526_1
|
Factor: E2F-3:HES-7; motif: NNNSGCGCSNNNNNCRCGYGNN; match class: 1
|
0.000110099416798622
|
14997
|
90
|
85
|
0.944444444444444
|
0.00566780022671201
|
|
TF
|
TF:M00338
|
Factor: ATF; motif: NTGACGTCANYS
|
0.000110905860416078
|
3798
|
29
|
17
|
0.586206896551724
|
0.00447604002106372
|
|
TF
|
TF:M12153
|
Factor: Sp3; motif: NGCCACGCCCMCN
|
0.000114538266349845
|
6205
|
82
|
46
|
0.560975609756098
|
0.00741337630942788
|
|
TF
|
TF:M01219
|
Factor: SP1:SP3; motif: CCSCCCCCYCC
|
0.000116051805802261
|
6812
|
89
|
52
|
0.584269662921348
|
0.00763358778625954
|
|
TF
|
TF:M04152_1
|
Factor: TFAP2C; motif: NGCCCNNRGGCA; match class: 1
|
0.000117596368787554
|
5580
|
82
|
43
|
0.524390243902439
|
0.00770609318996416
|
|
TF
|
TF:M04863
|
Factor: TF3C-beta; motif: CCNGGAGGGCTTCCTGGAGGAG
|
0.000124681325223177
|
13618
|
98
|
87
|
0.887755102040816
|
0.00638860331913644
|
|
TF
|
TF:M01862
|
Factor: ATF-2; motif: NNNTGACGTNAN
|
0.000124681325223177
|
3230
|
30
|
16
|
0.533333333333333
|
0.00495356037151703
|
|
TF
|
TF:M09723_1
|
Factor: BTEB1; motif: GGGGGCGGGGCNGSGGGNGS; match class: 1
|
0.000124681325223177
|
6113
|
88
|
48
|
0.545454545454545
|
0.00785211843611974
|
|
TF
|
TF:M12120_1
|
Factor: FPM315; motif: NGTGCTCCCN; match class: 1
|
0.000124681325223177
|
10
|
63
|
3
|
0.0476190476190476
|
0.3
|
|
TF
|
TF:M12057_1
|
Factor: ZBP99; motif: GCCCMTCCCCCR; match class: 1
|
0.00013089467950671
|
1359
|
63
|
16
|
0.253968253968254
|
0.0117733627667403
|
|
TF
|
TF:M12313_1
|
Factor: ZNF460; motif: NNACNCCCCCCNN; match class: 1
|
0.000132780127638594
|
2203
|
96
|
27
|
0.28125
|
0.0122560145256468
|
|
TF
|
TF:M02090
|
Factor: E2F-4; motif: GCGGGAAANA
|
0.000133105057100062
|
11966
|
96
|
79
|
0.822916666666667
|
0.00660203911081397
|
|
TF
|
TF:M12183_1
|
Factor: KLF14; motif: NRCCACRCCCMCN; match class: 1
|
0.000137805448434103
|
2487
|
96
|
29
|
0.302083333333333
|
0.0116606353035786
|
|
TF
|
TF:M11696_1
|
Factor: MEF-2B1; motif: CCNWATWWRG; match class: 1
|
0.000139301577553609
|
898
|
4
|
4
|
1
|
0.0044543429844098
|
|
TF
|
TF:M04823
|
Factor: E2F-4; motif: NNTTCCCGCCNN
|
0.000140027979341156
|
12747
|
92
|
79
|
0.858695652173913
|
0.00619753667529615
|
|
TF
|
TF:M07250
|
Factor: E2F-1; motif: NNNSSCGCSAANN
|
0.00014118274305424
|
10703
|
89
|
69
|
0.775280898876405
|
0.00644679061945249
|
|
TF
|
TF:M11337_1
|
Factor: ATF-4; motif: GGATGACGTCATCC; match class: 1
|
0.000147800127434672
|
2806
|
34
|
16
|
0.470588235294118
|
0.00570206699928724
|
|
TF
|
TF:M12183
|
Factor: KLF14; motif: NRCCACRCCCMCN
|
0.000147800127434672
|
6878
|
89
|
52
|
0.584269662921348
|
0.00756033730735679
|
|
TF
|
TF:M04797
|
Factor: Egr-1; motif: CCGCCCMCG
|
0.000147800127434672
|
5116
|
89
|
43
|
0.48314606741573
|
0.00840500390930414
|
|
TF
|
TF:M11601_1
|
Factor: TCF-1; motif: ACATCGRGRCGCTGW; match class: 1
|
0.000148791742603286
|
11053
|
90
|
71
|
0.788888888888889
|
0.00642359540396273
|
|
TF
|
TF:M09887
|
Factor: CREB1; motif: NRRTGACGTMA
|
0.000152703301254818
|
5419
|
72
|
38
|
0.527777777777778
|
0.00701236390477948
|
|
TF
|
TF:M00469
|
Factor: AP-2alpha; motif: GCCNNNRGS
|
0.000156026958666098
|
5147
|
86
|
42
|
0.488372093023256
|
0.00816009325820866
|
|
TF
|
TF:M02023
|
Factor: MAZ; motif: NKGGGAGGGGRGGR
|
0.000156545746908643
|
7134
|
95
|
56
|
0.589473684210526
|
0.00784973366975049
|
|
TF
|
TF:M08207_1
|
Factor: E2F-3:TBR2; motif: ANGTGYKANGGCGCSTTNNCRNNT; match class: 1
|
0.000164205951468208
|
15466
|
90
|
86
|
0.955555555555556
|
0.00556058450795293
|
|
TF
|
TF:M00177_1
|
Factor: CREB; motif: NSTGACGTAANN; match class: 1
|
0.000171141367608854
|
1363
|
79
|
18
|
0.227848101265823
|
0.0132061628760088
|
|
TF
|
TF:M01972
|
Factor: EGR-1; motif: TGCGTGGGCGK
|
0.000192526296717401
|
5360
|
89
|
44
|
0.49438202247191
|
0.0082089552238806
|
|
TF
|
TF:M00039
|
Factor: CREB; motif: TGACGTMA
|
0.000201524867772775
|
5222
|
83
|
41
|
0.493975903614458
|
0.00785139793182689
|
|
TF
|
TF:M01973
|
Factor: PLAG1; motif: CCCCCKWNNNGGSCCC
|
0.000209640725445207
|
6167
|
96
|
51
|
0.53125
|
0.00826982325279715
|
|
TF
|
TF:M11217
|
Factor: CREB1; motif: NRTGACGYN
|
0.000218728246860885
|
2783
|
98
|
31
|
0.316326530612245
|
0.0111390585698886
|
|
TF
|
TF:M12157
|
Factor: KLF15; motif: RCCACGCCCCCC
|
0.000222957568969213
|
3619
|
96
|
36
|
0.375
|
0.00994749930920144
|
|
TF
|
TF:M07436
|
Factor: WT1; motif: NNGGGNGGGSGN
|
0.000222957568969213
|
6182
|
96
|
51
|
0.53125
|
0.00824975736007764
|
|
TF
|
TF:M00917
|
Factor: CREB; motif: CNNTGACGTMA
|
0.000222957568969213
|
5578
|
84
|
43
|
0.511904761904762
|
0.00770885622086769
|
|
TF
|
TF:M05444_1
|
Factor: CPBP; motif: NGGGCGG; match class: 1
|
0.000222957568969213
|
3313
|
88
|
32
|
0.363636363636364
|
0.00965891940839119
|
|
TF
|
TF:M09866_1
|
Factor: ATF-1; motif: NNRTGACGYMA; match class: 1
|
0.000222957568969213
|
955
|
72
|
14
|
0.194444444444444
|
0.0146596858638743
|
|
TF
|
TF:M05361_1
|
Factor: Sp6; motif: WGGGCGG; match class: 1
|
0.000222957568969213
|
3313
|
88
|
32
|
0.363636363636364
|
0.00965891940839119
|
|
TF
|
TF:M11335_1
|
Factor: ATF-4; motif: GGATGACGTCATCC; match class: 1
|
0.000222957568969213
|
2207
|
57
|
19
|
0.333333333333333
|
0.00860897145446307
|
|
TF
|
TF:M05332_1
|
Factor: Sp2; motif: WGGGCGG; match class: 1
|
0.000222957568969213
|
3313
|
88
|
32
|
0.363636363636364
|
0.00965891940839119
|
|
TF
|
TF:M11337
|
Factor: ATF-4; motif: GGATGACGTCATCC
|
0.000222957568969213
|
2911
|
34
|
16
|
0.470588235294118
|
0.00549639299209894
|
|
TF
|
TF:M03922
|
Factor: SP4; motif: NWRGCCACGCCCMCTYN
|
0.000230386759287728
|
6046
|
98
|
51
|
0.520408163265306
|
0.0084353291432352
|
|
TF
|
TF:M00428
|
Factor: E2F-1; motif: NKTSSCGC
|
0.000236123706679573
|
8947
|
89
|
61
|
0.685393258426966
|
0.00681792779702694
|
|
TF
|
TF:M09600_1
|
Factor: CREB1; motif: TKACGTCAYNN; match class: 1
|
0.000236458916028504
|
1552
|
72
|
18
|
0.25
|
0.0115979381443299
|
|
TF
|
TF:M10076_1
|
Factor: SRF; motif: TTNCCWTWTWTGGNCWNN; match class: 1
|
0.000238688057982283
|
12
|
68
|
3
|
0.0441176470588235
|
0.25
|
|
TF
|
TF:M04516
|
Factor: E2F-1; motif: TTTGGCGCCAAA
|
0.000238688057982283
|
11383
|
86
|
69
|
0.802325581395349
|
0.00606167091276465
|
|
TF
|
TF:M11285_1
|
Factor: Fra-1; motif: GRTGACGTCATC; match class: 1
|
0.000250768875069539
|
4589
|
29
|
18
|
0.620689655172414
|
0.00392242318587928
|
|
TF
|
TF:M09898
|
Factor: Egr-2; motif: GNGRRNGWGKGGGNGGRG
|
0.000250813987864609
|
6484
|
95
|
52
|
0.547368421052632
|
0.00801974090067859
|
|
TF
|
TF:M09600
|
Factor: CREB1; motif: TKACGTCAYNN
|
0.000258803417352883
|
5324
|
72
|
37
|
0.513888888888889
|
0.00694966190833959
|
|
TF
|
TF:M11401
|
Factor: Fli-1; motif: NACCGGATATCCGGTN
|
0.000260160023007331
|
11952
|
86
|
71
|
0.825581395348837
|
0.00594042838018742
|
|
TF
|
TF:M11697
|
Factor: MEF-2B1; motif: CCNWATWNRG
|
0.0002623332546977
|
1096
|
4
|
4
|
1
|
0.00364963503649635
|
|
TF
|
TF:M09868
|
Factor: ATF-3; motif: GGTSACGTGAN
|
0.0002623332546977
|
6391
|
87
|
48
|
0.551724137931034
|
0.0075105617274292
|
|
TF
|
TF:M10098
|
Factor: DP-1; motif: NRNNGGCGGGAANN
|
0.000277858752748374
|
7015
|
85
|
50
|
0.588235294117647
|
0.00712758374910905
|
|
TF
|
TF:M00017_1
|
Factor: ATF; motif: CNSTGACGTNNNYC; match class: 1
|
0.000286207539682952
|
2222
|
73
|
22
|
0.301369863013699
|
0.0099009900990099
|
|
TF
|
TF:M00186_1
|
Factor: SRF; motif: GNCCAWATAWGGMN; match class: 1
|
0.000297309999569486
|
13
|
68
|
3
|
0.0441176470588235
|
0.230769230769231
|
|
TF
|
TF:M00179
|
Factor: ATF2; motif: VGTGACGTMACN
|
0.000297528567564095
|
6065
|
84
|
45
|
0.535714285714286
|
0.00741962077493817
|
|
TF
|
TF:M09868_1
|
Factor: ATF-3; motif: GGTSACGTGAN; match class: 1
|
0.000297528567564095
|
2116
|
82
|
23
|
0.280487804878049
|
0.0108695652173913
|
|
TF
|
TF:M11530_1
|
Factor: E2F-2; motif: NWTTTGGCGCCAWWNN; match class: 1
|
0.000306933724676231
|
13449
|
86
|
76
|
0.883720930232558
|
0.00565097776786378
|
|
TF
|
TF:M01865
|
Factor: BTEB3; motif: BNRNGGGAGGNGT
|
0.000323938541770641
|
9899
|
96
|
69
|
0.71875
|
0.00697040105061117
|
|
TF
|
TF:M11695
|
Factor: MEF-2D; motif: CCAWATWTRG
|
0.000324343963847929
|
288
|
4
|
3
|
0.75
|
0.0104166666666667
|
|
TF
|
TF:M11393
|
Factor: Erg; motif: NACCGGATATCCGGTN
|
0.000341833395997286
|
14105
|
86
|
78
|
0.906976744186046
|
0.00552995391705069
|
|
TF
|
TF:M11335
|
Factor: ATF-4; motif: GGATGACGTCATCC
|
0.000341833395997286
|
2292
|
57
|
19
|
0.333333333333333
|
0.00828970331588133
|
|
TF
|
TF:M07289
|
Factor: GKLF; motif: NNNRGGNGNGGSN
|
0.00034435370062597
|
14646
|
90
|
83
|
0.922222222222222
|
0.00566707633483545
|
|
TF
|
TF:M01299
|
Factor: MECP2; motif: CCGGNNTTWA
|
0.00034435370062597
|
5228
|
93
|
44
|
0.473118279569892
|
0.00841622035195103
|
|
TF
|
TF:M01972_1
|
Factor: EGR-1; motif: TGCGTGGGCGK; match class: 1
|
0.000349947751320057
|
1472
|
124
|
24
|
0.193548387096774
|
0.016304347826087
|
|
TF
|
TF:M11309_1
|
Factor: ATF-2; motif: NRTGACGTMANN; match class: 1
|
0.000351715653945062
|
2293
|
72
|
22
|
0.305555555555556
|
0.00959441779328391
|
|
TF
|
TF:M01857_1
|
Factor: AP-2alpha; motif: NGCCYSNNGSN; match class: 1
|
0.000352706062897774
|
4616
|
110
|
46
|
0.418181818181818
|
0.00996533795493934
|
|
TF
|
TF:M11217_1
|
Factor: CREB1; motif: NRTGACGYN; match class: 1
|
0.000359042660156684
|
457
|
79
|
10
|
0.126582278481013
|
0.0218818380743982
|
|
TF
|
TF:M11401_1
|
Factor: Fli-1; motif: NACCGGATATCCGGTN; match class: 1
|
0.000376783963126836
|
11801
|
93
|
75
|
0.806451612903226
|
0.00635539361071096
|
|
TF
|
TF:M07611_1
|
Factor: p54NRB; motif: GRNNNMGGATGRMNNCGGA; match class: 1
|
0.000384800084344194
|
1645
|
12
|
7
|
0.583333333333333
|
0.00425531914893617
|
|
TF
|
TF:M01299_1
|
Factor: MECP2; motif: CCGGNNTTWA; match class: 1
|
0.000396975449091545
|
1032
|
90
|
16
|
0.177777777777778
|
0.0155038759689922
|
|
TF
|
TF:M11302
|
Factor: batf3; motif: GATGAYGTCATN
|
0.000410922095256814
|
308
|
72
|
8
|
0.111111111111111
|
0.025974025974026
|
|
TF
|
TF:M11603
|
Factor: TCF-1; motif: ACATCGRGRCGCTGW
|
0.000419129981394003
|
16361
|
98
|
95
|
0.969387755102041
|
0.0058064910457796
|
|
TF
|
TF:M00483
|
Factor: ATF6; motif: TGACGTGG
|
0.000422243040319549
|
2627
|
83
|
26
|
0.313253012048193
|
0.0098972211648268
|
|
TF
|
TF:M09603
|
Factor: Egr-1; motif: NNNNNGYGKGGGNGGGNN
|
0.000424062752563502
|
4627
|
95
|
41
|
0.431578947368421
|
0.00886103306678193
|
|
TF
|
TF:M08209
|
Factor: E2F-3:FOXO6; motif: NAATGACACGCGCCCMC
|
0.000434223157647771
|
13673
|
83
|
74
|
0.891566265060241
|
0.00541212608791048
|
|
TF
|
TF:M06948_1
|
Factor: Sp2; motif: TGGGCGCGCCCA; match class: 1
|
0.000442100253025355
|
6205
|
98
|
51
|
0.520408163265306
|
0.00821917808219178
|
|
TF
|
TF:M07218
|
Factor: MEF-2A; motif: NKCTAAAAATAGMNN
|
0.000452828283947093
|
311
|
11
|
4
|
0.363636363636364
|
0.0128617363344051
|
|
TF
|
TF:M00736_1
|
Factor: E2F-1:DP-1; motif: TTTCSCGC; match class: 1
|
0.000461685891386126
|
4639
|
89
|
39
|
0.438202247191011
|
0.00840698426384997
|
|
TF
|
TF:M03971_1
|
Factor: ELK1; motif: NACTTCCGSCGGAAGYN; match class: 1
|
0.000465853234836609
|
7157
|
85
|
50
|
0.588235294117647
|
0.00698616738857063
|
|
TF
|
TF:M03567_1
|
Factor: Sp2; motif: NYSGCCCCGCCCCCY; match class: 1
|
0.000486589468070534
|
3614
|
96
|
35
|
0.364583333333333
|
0.00968456004427228
|
|
TF
|
TF:M12151
|
Factor: Sp1; motif: NWRGCCMCGCCCMCN
|
0.000495003723140919
|
6821
|
82
|
47
|
0.573170731707317
|
0.00689048526609002
|
|
TF
|
TF:M12345_1
|
Factor: Zbtb37; motif: NYACCGCRNTCACCGCR; match class: 1
|
0.000495003723140919
|
1688
|
84
|
20
|
0.238095238095238
|
0.0118483412322275
|
|
TF
|
TF:M03989_1
|
Factor: FLI1; motif: ACCGGAAATCCGGT; match class: 1
|
0.000495003723140919
|
5595
|
89
|
44
|
0.49438202247191
|
0.00786416443252904
|
|
TF
|
TF:M10107
|
Factor: DB1; motif: GGRRRRGRRGGAGGGGGNGRRR
|
0.000495003723140919
|
5822
|
59
|
33
|
0.559322033898305
|
0.00566815527310203
|
|
TF
|
TF:M00915
|
Factor: AP-2; motif: SNNNCCNCAGGCN
|
0.000523395596092039
|
9612
|
101
|
70
|
0.693069306930693
|
0.00728256346233874
|
|
TF
|
TF:M11395_1
|
Factor: Erg; motif: NACCGGAWWTCCGGTN; match class: 1
|
0.000550268795926222
|
7963
|
92
|
57
|
0.619565217391304
|
0.00715810624136632
|
|
TF
|
TF:M10426
|
Factor: ctcf; motif: CCRSCAGGGGGCGCN
|
0.000588343563387687
|
9305
|
90
|
62
|
0.688888888888889
|
0.00666308436324557
|
|
TF
|
TF:M00915_1
|
Factor: AP-2; motif: SNNNCCNCAGGCN; match class: 1
|
0.000588343563387687
|
4551
|
104
|
43
|
0.413461538461538
|
0.00944847286310701
|
|
TF
|
TF:M07615
|
Factor: Sp3; motif: GGGGCGGGGSNN
|
0.000593891445654073
|
6807
|
96
|
53
|
0.552083333333333
|
0.0077861025415014
|
|
TF
|
TF:M01199_1
|
Factor: RNF96; motif: BCCCGCRGCC; match class: 1
|
0.000599598415453827
|
4322
|
86
|
36
|
0.418604651162791
|
0.00832947709393799
|
|
TF
|
TF:M00916_1
|
Factor: CREB; motif: NNTKACGTCANNNS; match class: 1
|
0.000626947183902975
|
1692
|
72
|
18
|
0.25
|
0.0106382978723404
|
|
TF
|
TF:M00695_1
|
Factor: ETF; motif: GVGGMGG; match class: 1
|
0.000634729792234668
|
7032
|
98
|
55
|
0.561224489795918
|
0.00782138794084187
|
|
TF
|
TF:M12227
|
Factor: ZIC4; motif: NNCCNCCCRYNGYGN
|
0.000641527928157759
|
11063
|
88
|
68
|
0.772727272727273
|
0.00614661484226702
|
|
TF
|
TF:M08767_1
|
Factor: HES-1; motif: GNCACGTGNC; match class: 1
|
0.00067150309204838
|
7709
|
95
|
57
|
0.6
|
0.00739395511739525
|
|
TF
|
TF:M11309
|
Factor: ATF-2; motif: NRTGACGTMANN
|
0.00068498485307722
|
6429
|
34
|
23
|
0.676470588235294
|
0.00357753927515943
|
|
TF
|
TF:M07289_1
|
Factor: GKLF; motif: NNNRGGNGNGGSN; match class: 1
|
0.000689039631300417
|
10555
|
96
|
71
|
0.739583333333333
|
0.00672666982472762
|
|
TF
|
TF:M11022
|
Factor: IRX2a; motif: ACRYGNNNNACRYGT
|
0.000692745362441218
|
9487
|
89
|
62
|
0.696629213483146
|
0.00653525877516602
|
|
TF
|
TF:M04025_1
|
Factor: SRF; motif: TGMCCATATATGGKCA; match class: 1
|
0.000702316131729005
|
18
|
68
|
3
|
0.0441176470588235
|
0.166666666666667
|
|
TF
|
TF:M11061
|
Factor: HES-1; motif: GGCRCGTGNC
|
0.00070324701821586
|
6271
|
89
|
47
|
0.528089887640449
|
0.00749481741349067
|
|
TF
|
TF:M00446
|
Factor: Spz1; motif: DNNGGRGGGWWNNNN
|
0.00070946551285484
|
8655
|
31
|
25
|
0.806451612903226
|
0.00288850375505488
|
|
TF
|
TF:M04934_1
|
Factor: TR4; motif: ACCCCGS; match class: 1
|
0.000760489004504085
|
15478
|
89
|
84
|
0.943820224719101
|
0.00542705775940044
|
|
TF
|
TF:M04152
|
Factor: TFAP2C; motif: NGCCCNNRGGCA
|
0.000762294982006416
|
7738
|
89
|
54
|
0.606741573033708
|
0.00697854742827604
|
|
TF
|
TF:M04516_1
|
Factor: E2F-1; motif: TTTGGCGCCAAA; match class: 1
|
0.000774979675893968
|
10227
|
89
|
65
|
0.730337078651685
|
0.00635572504155666
|
|
TF
|
TF:M01861_1
|
Factor: ATF-1; motif: TNACGTCAN; match class: 1
|
0.000774979675893968
|
1896
|
72
|
19
|
0.263888888888889
|
0.0100210970464135
|
|
TF
|
TF:M11100
|
Factor: Max; motif: NNCACGTGYN
|
0.000774979675893968
|
82
|
89
|
5
|
0.0561797752808989
|
0.0609756097560976
|
|
TF
|
TF:M07206
|
Factor: E2F-1; motif: NGGGCGGGARV
|
0.000806215799589052
|
14575
|
96
|
87
|
0.90625
|
0.00596912521440823
|
|
TF
|
TF:M11081
|
Factor: SREBP-1; motif: RTCRCGTGAY
|
0.000820756990366455
|
11062
|
35
|
31
|
0.885714285714286
|
0.00280238654854457
|
|
TF
|
TF:M09892
|
Factor: E2F-1; motif: NNNNGGCGGGAARN
|
0.000822801355816225
|
13528
|
92
|
80
|
0.869565217391304
|
0.00591366055588409
|
|
TF
|
TF:M09895
|
Factor: E2F-6; motif: NGGGCGGGARRNN
|
0.000827181102709345
|
7765
|
91
|
55
|
0.604395604395604
|
0.00708306503541533
|
|
TF
|
TF:M00806
|
Factor: NF-1; motif: NTGGNNNNNNGCCAANN
|
0.000828386769334241
|
4297
|
101
|
40
|
0.396039603960396
|
0.00930882010705143
|
|
TF
|
TF:M01843
|
Factor: DEC2; motif: SNTCACGTGS
|
0.000836348926785019
|
3788
|
66
|
27
|
0.409090909090909
|
0.00712777191129884
|
|
TF
|
TF:M03807_1
|
Factor: SP2; motif: GNNGGGGGCGGGGSN; match class: 1
|
0.000848962495407388
|
4199
|
124
|
46
|
0.370967741935484
|
0.0109549892831627
|
|
TF
|
TF:M00189
|
Factor: AP-2; motif: MKCCCSCNGGCG
|
0.000848962495407388
|
10621
|
96
|
71
|
0.739583333333333
|
0.00668486959796629
|
|
TF
|
TF:M07436_1
|
Factor: WT1; motif: NNGGGNGGGSGN; match class: 1
|
0.000848962495407388
|
2405
|
94
|
26
|
0.276595744680851
|
0.0108108108108108
|
|
TF
|
TF:M08911_1
|
Factor: CTCF; motif: NCCRSTAGGGGGCGC; match class: 1
|
0.000850775144956904
|
3934
|
88
|
34
|
0.386363636363636
|
0.00864260294865277
|
|
TF
|
TF:M04110
|
Factor: RUNX3; motif: NRACCGCANWAACCRCAN
|
0.000860654480115493
|
9266
|
86
|
59
|
0.686046511627907
|
0.00636736455860134
|
|
TF
|
TF:M00694
|
Factor: E4F1; motif: SYTACGTCAC
|
0.000860654480115493
|
152
|
120
|
7
|
0.0583333333333333
|
0.0460526315789474
|
|
TF
|
TF:M07261_1
|
Factor: LKLF; motif: GGGGTGGKSN; match class: 1
|
0.000870523005917178
|
4062
|
96
|
37
|
0.385416666666667
|
0.00910881339241753
|
|
TF
|
TF:M10435
|
Factor: Sp2; motif: GGGGCGGGG
|
0.000884374560508881
|
7696
|
96
|
57
|
0.59375
|
0.00740644490644491
|
|
TF
|
TF:M00917_1
|
Factor: CREB; motif: CNNTGACGTMA; match class: 1
|
0.000884374560508881
|
1590
|
4
|
4
|
1
|
0.00251572327044025
|
|
TF
|
TF:M07063
|
Factor: Sp1; motif: GGGGCGGGGC
|
0.000884374560508881
|
7696
|
96
|
57
|
0.59375
|
0.00740644490644491
|
|
TF
|
TF:M00922_1
|
Factor: SRF; motif: CCAWATAWGGMNMNG; match class: 1
|
0.000897178183694973
|
20
|
68
|
3
|
0.0441176470588235
|
0.15
|
|
TF
|
TF:M08209_1
|
Factor: E2F-3:FOXO6; motif: NAATGACACGCGCCCMC; match class: 1
|
0.000910297057135866
|
7798
|
89
|
54
|
0.606741573033708
|
0.00692485252628879
|
|
TF
|
TF:M00430
|
Factor: E2F-1; motif: NTTSGCGG
|
0.00091806035986667
|
8016
|
89
|
55
|
0.617977528089888
|
0.00686127744510978
|
|
TF
|
TF:M04107_1
|
Factor: RUNX2; motif: WRACCGCANWAACCGCAN; match class: 1
|
0.000920462724769744
|
6444
|
95
|
50
|
0.526315789473684
|
0.00775915580384854
|
|
TF
|
TF:M07261
|
Factor: LKLF; motif: GGGGTGGKSN
|
0.000925215203558411
|
9774
|
96
|
67
|
0.697916666666667
|
0.0068549212195621
|
|
TF
|
TF:M11881_1
|
Factor: pax-6; motif: NYACGCNTSANYGCNYN; match class: 1
|
0.000930875129292309
|
7158
|
87
|
50
|
0.574712643678161
|
0.0069851913942442
|
|
TF
|
TF:M08767
|
Factor: HES-1; motif: GNCACGTGNC
|
0.00093505880526839
|
7813
|
95
|
57
|
0.6
|
0.0072955330858825
|
|
TF
|
TF:M04106_1
|
Factor: RUNX2; motif: NRACCGCAAACCGCAN; match class: 1
|
0.000942926355103803
|
8372
|
86
|
55
|
0.63953488372093
|
0.00656951743908266
|
|
TF
|
TF:M08205_1
|
Factor: E2F-1:Elk-1; motif: SGCGCSNNAMCGGAAGT; match class: 1
|
0.000943964017993911
|
10438
|
96
|
70
|
0.729166666666667
|
0.0067062655681165
|
|
TF
|
TF:M09969
|
Factor: EKLF; motif: NGGGYGKGGCNNGG
|
0.000944269314004758
|
4589
|
96
|
40
|
0.416666666666667
|
0.00871649596862061
|
|
TF
|
TF:M12057
|
Factor: ZBP99; motif: GCCCMTCCCCCR
|
0.000964573642768683
|
5225
|
83
|
39
|
0.469879518072289
|
0.00746411483253588
|
|
TF
|
TF:M00425
|
Factor: E2F; motif: TTTCGCGC
|
0.000989898257293135
|
11043
|
89
|
68
|
0.764044943820225
|
0.00615774698904283
|
|
TF
|
TF:M00179_1
|
Factor: ATF2; motif: VGTGACGTMACN; match class: 1
|
0.00104107845152408
|
1778
|
72
|
18
|
0.25
|
0.0101237345331834
|
|
TF
|
TF:M11291_1
|
Factor: ATF-3; motif: NRTGACGTCAYN; match class: 1
|
0.00105764248442752
|
4408
|
20
|
13
|
0.65
|
0.0029491833030853
|
|
TF
|
TF:M11877_1
|
Factor: pax-2; motif: NCGTCACGCNYSRNYGCNYN; match class: 1
|
0.00106709547690105
|
5304
|
24
|
16
|
0.666666666666667
|
0.00301659125188537
|
|
TF
|
TF:M10071_1
|
Factor: Sp1; motif: NGGGGGCGGGGCCNGGGGGGGG; match class: 1
|
0.00110141511509619
|
4623
|
96
|
40
|
0.416666666666667
|
0.00865239022279905
|
|
TF
|
TF:M04146_1
|
Factor: TFAP2A; motif: YGCCCNNRGGCN; match class: 1
|
0.00110141511509619
|
3800
|
74
|
29
|
0.391891891891892
|
0.00763157894736842
|
|
TF
|
TF:M09973_1
|
Factor: CPBP; motif: GNNRGGGHGGGGNNGGGRN; match class: 1
|
0.00112412925444476
|
6607
|
96
|
51
|
0.53125
|
0.00771908581807174
|
|
TF
|
TF:M00513
|
Factor: ATF3; motif: CBCTGACGTCANCS
|
0.00113592280270951
|
6386
|
31
|
21
|
0.67741935483871
|
0.00328844347009082
|
|
TF
|
TF:M00322
|
Factor: c-Myc:Max; motif: GCCAYGYGSN
|
0.00115588607621785
|
7873
|
95
|
57
|
0.6
|
0.00723993395147974
|
|
TF
|
TF:M00177
|
Factor: CREB; motif: NSTGACGTAANN
|
0.00122636095735347
|
5221
|
98
|
44
|
0.448979591836735
|
0.00842750430951925
|
|
TF
|
TF:M09734_1
|
Factor: ZNF692; motif: SYNGGSCCCASCCNC; match class: 1
|
0.00122636095735347
|
6255
|
96
|
49
|
0.510416666666667
|
0.00783373301358913
|
|
TF
|
TF:M07129
|
Factor: Sp2; motif: GYCCCGCCYCYNNNN
|
0.00125050512998137
|
6260
|
96
|
49
|
0.510416666666667
|
0.00782747603833866
|
|
TF
|
TF:M07040
|
Factor: GKLF; motif: NNRRGRRNGNSNNN
|
0.00131301880431499
|
12705
|
101
|
83
|
0.821782178217822
|
0.00653286107831562
|
|
TF
|
TF:M02011
|
Factor: HES-1; motif: GSCACGMGMC
|
0.00149159914079927
|
4259
|
68
|
29
|
0.426470588235294
|
0.00680911011974642
|
|
TF
|
TF:M03814
|
Factor: BTEB2; motif: GNAGGGGGNGGGSSNN
|
0.00153141672891793
|
5124
|
86
|
39
|
0.453488372093023
|
0.00761124121779859
|
|
TF
|
TF:M00333
|
Factor: ZF5; motif: NRNGNGCGCGCWN
|
0.00153560664958849
|
15072
|
90
|
83
|
0.922222222222222
|
0.00550690021231422
|
|
TF
|
TF:M08487
|
Factor: GCMa:Erg; motif: ATGCGGGCGGAARKG
|
0.00153560664958849
|
13942
|
96
|
84
|
0.875
|
0.00602496055085354
|
|
TF
|
TF:M11397_1
|
Factor: Erg; motif: NACCGGATATCCGGTN; match class: 1
|
0.00153560664958849
|
11731
|
93
|
73
|
0.78494623655914
|
0.00622282840337567
|
|
TF
|
TF:M09967
|
Factor: AP-2rep; motif: NGGGGCGGGGC
|
0.00156182967209821
|
4023
|
96
|
36
|
0.375
|
0.00894854586129754
|
|
TF
|
TF:M09893
|
Factor: E2F-3; motif: NNGGCGGGAAA
|
0.00162846552684092
|
6065
|
82
|
42
|
0.51219512195122
|
0.00692497938994229
|
|
TF
|
TF:M12140
|
Factor: Egr-1; motif: NNCRCCCMCGCNN
|
0.00165741865247743
|
5360
|
83
|
39
|
0.469879518072289
|
0.00727611940298507
|
|
TF
|
TF:M11066
|
Factor: HES-7; motif: GNCACGYGNN
|
0.00166950842715127
|
11507
|
96
|
74
|
0.770833333333333
|
0.00643086816720257
|
|
TF
|
TF:M03920
|
Factor: SP1; motif: RCCMCRCCCMC
|
0.00166950842715127
|
7283
|
96
|
54
|
0.5625
|
0.00741452698063985
|
|
TF
|
TF:M11220_1
|
Factor: CREB1; motif: NNTGACGTCANN; match class: 1
|
0.00168395841291716
|
1713
|
34
|
11
|
0.323529411764706
|
0.00642148277875073
|
|
TF
|
TF:M11397
|
Factor: Erg; motif: NACCGGATATCCGGTN
|
0.00168998815080118
|
12005
|
93
|
74
|
0.795698924731183
|
0.00616409829237818
|
|
TF
|
TF:M07084
|
Factor: E2F-4; motif: NGGCGGGAARN
|
0.00171822892204288
|
10834
|
96
|
71
|
0.739583333333333
|
0.00655344286505446
|
|
TF
|
TF:M11530
|
Factor: E2F-2; motif: NWTTTGGCGCCAWWNN
|
0.00173614257159132
|
14593
|
86
|
78
|
0.906976744186046
|
0.00534502843829233
|
|
TF
|
TF:M00691
|
Factor: ATF-1; motif: CYYTGACGTCA
|
0.00176903162045859
|
4489
|
23
|
14
|
0.608695652173913
|
0.00311873468478503
|
|
TF
|
TF:M11220
|
Factor: CREB1; motif: NNTGACGTCANN
|
0.00176903162045859
|
1725
|
34
|
11
|
0.323529411764706
|
0.0063768115942029
|
|
TF
|
TF:M08208_1
|
Factor: E2F-3:FOXI1; motif: NGACACCGCGCCCAC; match class: 1
|
0.00184133130409667
|
7127
|
22
|
17
|
0.772727272727273
|
0.00238529535568963
|
|
TF
|
TF:M04109
|
Factor: RUNX3; motif: NRACCGCAAACCGCAN
|
0.00186974604176103
|
9770
|
53
|
40
|
0.754716981132076
|
0.00409416581371546
|
|
TF
|
TF:M01100
|
Factor: LRF; motif: GGGGKYNNB
|
0.00186994161430635
|
7557
|
83
|
49
|
0.590361445783133
|
0.00648405451898902
|
|
TF
|
TF:M02012
|
Factor: HIF-1alpha; motif: NCACGT
|
0.00187982714319365
|
6648
|
25
|
18
|
0.72
|
0.00270758122743682
|
|
TF
|
TF:M00938
|
Factor: E2F-1; motif: TTGGCGCGRAANNGNM
|
0.00187982714319365
|
7334
|
98
|
55
|
0.561224489795918
|
0.00749931824379602
|
|
TF
|
TF:M09761_1
|
Factor: EGR1; motif: NGNGKGGGYGGNGS; match class: 1
|
0.00195135678636051
|
1568
|
124
|
23
|
0.185483870967742
|
0.0146683673469388
|
|
TF
|
TF:M11291
|
Factor: ATF-3; motif: NRTGACGTCAYN
|
0.00204477608433788
|
4720
|
20
|
13
|
0.65
|
0.00275423728813559
|
|
TF
|
TF:M00809
|
Factor: FOX; motif: KWTTGTTTRTTTW
|
0.00206678156298462
|
3746
|
21
|
12
|
0.571428571428571
|
0.00320341697810998
|
|
TF
|
TF:M00113_1
|
Factor: CREB; motif: NNGNTGACGTNN; match class: 1
|
0.00214526340074422
|
869
|
19
|
6
|
0.315789473684211
|
0.00690448791714614
|
|
TF
|
TF:M04744
|
Factor: ATF-3; motif: GGCGCSSNSNGRTSACGTSA
|
0.00215945414612505
|
10089
|
84
|
60
|
0.714285714285714
|
0.00594707106749926
|
|
TF
|
TF:M04025
|
Factor: SRF; motif: TGMCCATATATGGKCA
|
0.00216249703380299
|
28
|
68
|
3
|
0.0441176470588235
|
0.107142857142857
|
|
TF
|
TF:M09897_1
|
Factor: Egr-1; motif: NNNNGCGKGGGYGGNRN; match class: 1
|
0.00216806621956048
|
993
|
88
|
14
|
0.159090909090909
|
0.014098690835851
|
|
TF
|
TF:M00800_1
|
Factor: AP-2; motif: GSCCSCRGGCNRNRNN; match class: 1
|
0.00218663361096903
|
4268
|
96
|
37
|
0.385416666666667
|
0.00866916588566073
|
|
TF
|
TF:M11528_1
|
Factor: E2F-2; motif: NTTTTGGCGCCAWWWN; match class: 1
|
0.00219228299830203
|
4778
|
93
|
39
|
0.419354838709677
|
0.00816241105064881
|
|
TF
|
TF:M00736
|
Factor: E2F-1:DP-1; motif: TTTCSCGC
|
0.0022439503306655
|
10390
|
95
|
68
|
0.715789473684211
|
0.00654475457170356
|
|
TF
|
TF:M00807
|
Factor: Egr; motif: GTGGGSGCRRS
|
0.0022439503306655
|
4343
|
98
|
38
|
0.387755102040816
|
0.00874971218052038
|
|
TF
|
TF:M11880_1
|
Factor: pax-6; motif: NYACGCWTSANYGMNCN; match class: 1
|
0.00226881183978187
|
7046
|
82
|
46
|
0.560975609756098
|
0.00652852682372978
|
|
TF
|
TF:M03978_1
|
Factor: ETS1; motif: GCCGGAWGTACTTCCGGN; match class: 1
|
0.00227900420714962
|
6486
|
88
|
46
|
0.522727272727273
|
0.00709219858156028
|
|
TF
|
TF:M00144
|
Factor: Pax-5; motif: RRMSWGANWYCTNRAGCGKRACSRYNSM
|
0.00227900420714962
|
13205
|
82
|
70
|
0.853658536585366
|
0.00530102234002272
|
|
TF
|
TF:M00322_1
|
Factor: c-Myc:Max; motif: GCCAYGYGSN; match class: 1
|
0.00231959800767598
|
2862
|
124
|
34
|
0.274193548387097
|
0.0118798043326345
|
|
TF
|
TF:M01864_1
|
Factor: ATF-4; motif: KACGTCAKS; match class: 1
|
0.00232411569380896
|
1239
|
72
|
14
|
0.194444444444444
|
0.0112994350282486
|
|
TF
|
TF:M08867_1
|
Factor: AP2; motif: GCCYGSGGSN; match class: 1
|
0.00236924610578055
|
5186
|
84
|
38
|
0.452380952380952
|
0.00732741997686078
|
|
TF
|
TF:M09636
|
Factor: MAZ; motif: GGGMGGGGSSGGGGGGGGGGGG
|
0.00239373012838573
|
16244
|
101
|
96
|
0.95049504950495
|
0.00590987441516868
|
|
TF
|
TF:M01175
|
Factor: CKROX; motif: SCCCTCCCC
|
0.00239373012838573
|
7292
|
63
|
38
|
0.603174603174603
|
0.0052111903455842
|
|
TF
|
TF:M08662
|
Factor: AP-2gamma:HES-7; motif: NNCACGYGNNNNNNSCCNNNGGS
|
0.00240766220820018
|
1975
|
95
|
22
|
0.231578947368421
|
0.0111392405063291
|
|
TF
|
TF:M12182
|
Factor: BTEB3; motif: NRCCACGCCCMCN
|
0.00241266642579485
|
1738
|
79
|
18
|
0.227848101265823
|
0.0103567318757192
|
|
TF
|
TF:M04043
|
Factor: MYBL1; motif: ACCGTTAACSGY
|
0.00242088485075002
|
24
|
84
|
3
|
0.0357142857142857
|
0.125
|
|
TF
|
TF:M01588_1
|
Factor: GKLF; motif: GCCMCRCCCNNN; match class: 1
|
0.00243658479060509
|
3962
|
96
|
35
|
0.364583333333333
|
0.0088339222614841
|
|
TF
|
TF:M08329
|
Factor: GCMb:TBR2; motif: ATGCGGGNAGGTGTNA
|
0.00247336238821712
|
479
|
12
|
4
|
0.333333333333333
|
0.00835073068893528
|
|
TF
|
TF:M00426_1
|
Factor: E2F; motif: TTTSGCGS; match class: 1
|
0.00247336238821712
|
5830
|
71
|
36
|
0.507042253521127
|
0.00617495711835334
|
|
TF
|
TF:M11313
|
Factor: CREBPA; motif: NRTGACGTMANN
|
0.00252297346363529
|
3680
|
34
|
16
|
0.470588235294118
|
0.00434782608695652
|
|
TF
|
TF:M04863_1
|
Factor: TF3C-beta; motif: CCNGGAGGGCTTCCTGGAGGAG; match class: 1
|
0.00253697194669938
|
8573
|
85
|
54
|
0.635294117647059
|
0.00629884521171119
|
|
TF
|
TF:M09890_1
|
Factor: ctcf; motif: YGGCCACCAGRKGGCRSYN; match class: 1
|
0.00256048227247745
|
2884
|
34
|
14
|
0.411764705882353
|
0.00485436893203883
|
|
TF
|
TF:M11273
|
Factor: JunB; motif: GATGACGTCAYC
|
0.00256048227247745
|
9017
|
27
|
22
|
0.814814814814815
|
0.00243983586558722
|
|
TF
|
TF:M11266
|
Factor: c-Jun; motif: NATGACGTCAYN
|
0.00269516431580925
|
8688
|
31
|
24
|
0.774193548387097
|
0.00276243093922652
|
|
TF
|
TF:M10086_1
|
Factor: TAFII250; motif: RARRWGGCGGMGGNGR; match class: 1
|
0.00269516431580925
|
4153
|
96
|
36
|
0.375
|
0.00866843245846376
|
|
TF
|
TF:M01240
|
Factor: BEN; motif: CAGCGRNV
|
0.00270901328310663
|
16498
|
98
|
94
|
0.959183673469388
|
0.00569766032246333
|
|
TF
|
TF:M03976
|
Factor: ERG; motif: ACCGGAWATCCGGT
|
0.00272085547124021
|
12803
|
99
|
81
|
0.818181818181818
|
0.00632664219323596
|
|
TF
|
TF:M09970_1
|
Factor: KLF3; motif: NNNNNNGGGCGGGGCNNGN; match class: 1
|
0.00276074039295276
|
3783
|
82
|
30
|
0.365853658536585
|
0.00793021411578113
|
|
TF
|
TF:M11298
|
Factor: jdp2; motif: NRTGACGTCAYN
|
0.00276156151124758
|
1597
|
72
|
16
|
0.222222222222222
|
0.0100187852222918
|
|
TF
|
TF:M12152
|
Factor: Sp1; motif: NWRGCCACGCCCMCN
|
0.00276156151124758
|
7341
|
82
|
47
|
0.573170731707317
|
0.00640239749352949
|
|
TF
|
TF:M01783_1
|
Factor: SP2; motif: GGGCGGGAC; match class: 1
|
0.00279598582362659
|
4333
|
96
|
37
|
0.385416666666667
|
0.00853911839372259
|
|
TF
|
TF:M01303_1
|
Factor: SP1; motif: GGGGYGGGGNS; match class: 1
|
0.00284543648980999
|
3507
|
96
|
32
|
0.333333333333333
|
0.00912460792700314
|
|
TF
|
TF:M11696
|
Factor: MEF-2B1; motif: CCNWATWWRG
|
0.00284543648980999
|
2268
|
4
|
4
|
1
|
0.0017636684303351
|
|
TF
|
TF:M03932
|
Factor: ZIC4; motif: GRCCCCCCGCNGNGN
|
0.00284543648980999
|
2720
|
96
|
27
|
0.28125
|
0.00992647058823529
|
|
TF
|
TF:M08905
|
Factor: TIEG1; motif: GGSGGKGNNN
|
0.00284588993729754
|
5153
|
94
|
41
|
0.436170212765957
|
0.00795653017659616
|
|
TF
|
TF:M00115_1
|
Factor: Tax/CREB; motif: RTGACGCATAYCCCC; match class: 1
|
0.00286003055074425
|
2897
|
86
|
26
|
0.302325581395349
|
0.00897480151881257
|
|
TF
|
TF:M00791
|
Factor: HNF3; motif: NWRARYAAAYANN
|
0.00286003055074425
|
2732
|
32
|
13
|
0.40625
|
0.00475841874084919
|
|
TF
|
TF:M11215
|
Factor: N-Myc; motif: NNCACGTGNN
|
0.00289446839788389
|
1206
|
67
|
13
|
0.194029850746269
|
0.0107794361525705
|
|
TF
|
TF:M07384
|
Factor: HIF-1alpha; motif: NCACGTNN
|
0.00289446839788389
|
1206
|
67
|
13
|
0.194029850746269
|
0.0107794361525705
|
|
TF
|
TF:M03921
|
Factor: SP3; motif: NCCACGCCCMC
|
0.00294914192821324
|
3856
|
81
|
30
|
0.37037037037037
|
0.00778008298755187
|
|
TF
|
TF:M03557_1
|
Factor: P50; motif: GGRRANTCCCNN; match class: 1
|
0.00296652598546817
|
1292
|
63
|
13
|
0.206349206349206
|
0.010061919504644
|
|
TF
|
TF:M00338_1
|
Factor: ATF; motif: NTGACGTCANYS; match class: 1
|
0.00299301803276897
|
1122
|
72
|
13
|
0.180555555555556
|
0.0115864527629234
|
|
TF
|
TF:M00469_1
|
Factor: AP-2alpha; motif: GCCNNNRGS; match class: 1
|
0.0030570997387636
|
3334
|
68
|
24
|
0.352941176470588
|
0.00719856028794241
|
|
TF
|
TF:M08205
|
Factor: E2F-1:Elk-1; motif: SGCGCSNNAMCGGAAGT
|
0.00308276380625857
|
14429
|
96
|
85
|
0.885416666666667
|
0.00589091413126343
|
|
TF
|
TF:M03923
|
Factor: SP8; motif: RCCACGCCCMCY
|
0.00310958859620158
|
2843
|
74
|
23
|
0.310810810810811
|
0.00809004572634541
|
|
TF
|
TF:M05439_1
|
Factor: RREB-1; motif: GGGWCSA; match class: 1
|
0.00314668580251631
|
3200
|
123
|
36
|
0.292682926829268
|
0.01125
|
|
TF
|
TF:M10026_1
|
Factor: PATZ; motif: GGGGNGGGGGMKGGRRNGGNRN; match class: 1
|
0.00326271268990681
|
4894
|
96
|
40
|
0.416666666666667
|
0.0081732733959951
|
|
TF
|
TF:M00976
|
Factor: AhR,; motif: NRCGTGNGN
|
0.00326271268990681
|
4351
|
124
|
45
|
0.362903225806452
|
0.0103424500114916
|
|
TF
|
TF:M07461_1
|
Factor: KLF; motif: GGGNGGGG; match class: 1
|
0.0033216175701051
|
3397
|
104
|
33
|
0.317307692307692
|
0.00971445392993818
|
|
TF
|
TF:M11399_1
|
Factor: Fli-1; motif: NACCGGAWWTCCGGTY; match class: 1
|
0.0033216175701051
|
11513
|
93
|
71
|
0.763440860215054
|
0.00616694171805785
|
|
TF
|
TF:M11881
|
Factor: pax-6; motif: NYACGCNTSANYGCNYN
|
0.00336831060858102
|
13174
|
96
|
80
|
0.833333333333333
|
0.0060725671777744
|
|
TF
|
TF:M04918
|
Factor: Egr-1; motif: ACCGCCC
|
0.00339864872409442
|
5608
|
88
|
41
|
0.465909090909091
|
0.00731098430813124
|
|
TF
|
TF:M08207
|
Factor: E2F-3:TBR2; motif: ANGTGYKANGGCGCSTTNNCRNNT
|
0.00357145786135509
|
17055
|
90
|
88
|
0.977777777777778
|
0.00515977719143946
|
|
TF
|
TF:M00982_1
|
Factor: KROX; motif: CCCGCCCCCRCCCC; match class: 1
|
0.00357145786135509
|
3518
|
89
|
30
|
0.337078651685393
|
0.00852757248436612
|
|
TF
|
TF:M00243
|
Factor: Egr-1; motif: WTGCGTGGGCGK
|
0.00357145786135509
|
5356
|
89
|
40
|
0.449438202247191
|
0.00746825989544436
|
|
TF
|
TF:M01973_1
|
Factor: PLAG1; motif: CCCCCKWNNNGGSCCC; match class: 1
|
0.00358529651737286
|
2398
|
82
|
22
|
0.268292682926829
|
0.00917431192660551
|
|
TF
|
TF:M12217
|
Factor: GLIS2; motif: ACCCCCCRCGWNGC
|
0.00361218023691026
|
567
|
89
|
10
|
0.112359550561798
|
0.017636684303351
|
|
TF
|
TF:M08525
|
Factor: E2F-1:HES-7; motif: GGCRCGTGSYNNWNGGCGCSM
|
0.00369427606080547
|
17065
|
90
|
88
|
0.977777777777778
|
0.0051567535892177
|
|
TF
|
TF:M09862
|
Factor: ZNF614; motif: NCYCWGCCYYNNN
|
0.00376019746268824
|
9361
|
96
|
63
|
0.65625
|
0.00673005020831108
|
|
TF
|
TF:M00737
|
Factor: E2F-1:DP-2; motif: TTTSSCGC
|
0.00379901291825912
|
8771
|
79
|
51
|
0.645569620253165
|
0.00581461634933303
|
|
TF
|
TF:M01007
|
Factor: SRF; motif: CNKNKCCTTATWTGGNNNN
|
0.00381618264149934
|
256
|
68
|
6
|
0.0882352941176471
|
0.0234375
|
|
TF
|
TF:M10086
|
Factor: TAFII250; motif: RARRWGGCGGMGGNGR
|
0.00386408790277504
|
9262
|
88
|
58
|
0.659090909090909
|
0.00626214640466422
|
|
TF
|
TF:M00448
|
Factor: Zic1; motif: KGGGTGGTC
|
0.0038900491272532
|
10948
|
94
|
69
|
0.734042553191489
|
0.00630252100840336
|
|
TF
|
TF:M07246_1
|
Factor: ATF-4; motif: NNTKRCRTCANS; match class: 1
|
0.00400147460806815
|
584
|
72
|
9
|
0.125
|
0.0154109589041096
|
|
TF
|
TF:M12166
|
Factor: CPBP; motif: NGCCACGCCCN
|
0.00400850859141863
|
1328
|
89
|
16
|
0.179775280898876
|
0.0120481927710843
|
|
TF
|
TF:M07348
|
Factor: AP-2alpha; motif: NSCCNCRGGSN
|
0.00400850859141863
|
8047
|
83
|
50
|
0.602409638554217
|
0.00621349571268796
|
|
TF
|
TF:M11533_1
|
Factor: E2F-1; motif: NTTTTGGCGCCAWWWN; match class: 1
|
0.00403893315808048
|
8705
|
89
|
56
|
0.629213483146067
|
0.0064330844342332
|
|
TF
|
TF:M04595_1
|
Factor: SALL2; motif: GGGTGGG; match class: 1
|
0.004169465659738
|
7373
|
96
|
53
|
0.552083333333333
|
0.0071883900718839
|
|
TF
|
TF:M11307_1
|
Factor: ATF-2; motif: NNTGACGTMANN; match class: 1
|
0.004169465659738
|
1946
|
34
|
11
|
0.323529411764706
|
0.00565262076053443
|
|
TF
|
TF:M00514_1
|
Factor: ATF4; motif: CVTGACGYMABG; match class: 1
|
0.00421619753856696
|
1333
|
98
|
17
|
0.173469387755102
|
0.0127531882970743
|
|
TF
|
TF:M11541
|
Factor: Foxn2; motif: NNGCGTCNNNNNGACGCNN
|
0.00430560670376928
|
5751
|
87
|
41
|
0.471264367816092
|
0.00712919492262215
|
|
TF
|
TF:M08993
|
Factor: ZNF777; motif: GTCCGYCCCGTCSAACAAT
|
0.00435011587980852
|
10617
|
95
|
68
|
0.715789473684211
|
0.00640482245455402
|
|
TF
|
TF:M00244_1
|
Factor: NGFI-C; motif: WTGCGTGGGYGG; match class: 1
|
0.00435173115246808
|
1772
|
89
|
19
|
0.213483146067416
|
0.0107223476297968
|
|
TF
|
TF:M07246
|
Factor: ATF-4; motif: NNTKRCRTCANS
|
0.00438540021255176
|
2835
|
49
|
17
|
0.346938775510204
|
0.00599647266313933
|
|
TF
|
TF:M07380_1
|
Factor: E2F-4; motif: NTTTCSCGCC; match class: 1
|
0.0044231048158795
|
7965
|
82
|
49
|
0.597560975609756
|
0.0061519146264909
|
|
TF
|
TF:M07034_1
|
Factor: ATF-1; motif: NNNTGACGTNNN; match class: 1
|
0.00443242059492835
|
1179
|
72
|
13
|
0.180555555555556
|
0.0110262934690416
|
|
TF
|
TF:M01219_1
|
Factor: SP1:SP3; motif: CCSCCCCCYCC; match class: 1
|
0.00443242059492835
|
2937
|
126
|
34
|
0.26984126984127
|
0.0115764385427307
|
|
TF
|
TF:M00289
|
Factor: HFH3; motif: KNNTRTTTRTTTA
|
0.00443242059492835
|
2857
|
21
|
10
|
0.476190476190476
|
0.00350017500875044
|
|
TF
|
TF:M12154
|
Factor: Sp3; motif: NGCCACGCCCMCN
|
0.00443242059492835
|
2190
|
79
|
20
|
0.253164556962025
|
0.0091324200913242
|
|
TF
|
TF:M02090_1
|
Factor: E2F-4; motif: GCGGGAAANA; match class: 1
|
0.00448765642152413
|
6378
|
87
|
44
|
0.505747126436782
|
0.00689871433051113
|
|
TF
|
TF:M09775_1
|
Factor: MXI1; motif: NNNCCACGTGSNN; match class: 1
|
0.00453245167157211
|
39
|
67
|
3
|
0.0447761194029851
|
0.0769230769230769
|
|
TF
|
TF:M09896_1
|
Factor: E2F-7; motif: GRGGCGGGAANNN; match class: 1
|
0.00457401013973863
|
4525
|
85
|
34
|
0.4
|
0.00751381215469613
|
|
TF
|
TF:M11219
|
Factor: CREB1; motif: NRTGACGTR
|
0.00478521843722112
|
873
|
4
|
3
|
0.75
|
0.00343642611683849
|
|
TF
|
TF:M01597
|
Factor: Zfp281; motif: TGGGGGAGGGG
|
0.00480499347686881
|
1016
|
63
|
11
|
0.174603174603175
|
0.0108267716535433
|
|
TF
|
TF:M11478
|
Factor: AP-2beta; motif: NSCCNNNGGSN
|
0.00480994706051895
|
9942
|
87
|
60
|
0.689655172413793
|
0.00603500301750151
|
|
TF
|
TF:M05499_1
|
Factor: LKLF; motif: NGGGCGG; match class: 1
|
0.00483886816179489
|
2513
|
124
|
30
|
0.241935483870968
|
0.0119379228014326
|
|
TF
|
TF:M09645
|
Factor: Pax-5; motif: NNNNRNKCAGCNRAGCGTGAC
|
0.00483886816179489
|
2596
|
23
|
10
|
0.434782608695652
|
0.00385208012326656
|
|
TF
|
TF:M05386_1
|
Factor: KLF17; motif: NGGGCGG; match class: 1
|
0.00483886816179489
|
2513
|
124
|
30
|
0.241935483870968
|
0.0119379228014326
|
|
TF
|
TF:M00327_1
|
Factor: Pax-3; motif: NNNNNNCGTCACGSTYNNNNN; match class: 1
|
0.00485054642692361
|
6888
|
84
|
45
|
0.535714285714286
|
0.00653310104529617
|
|
TF
|
TF:M11327
|
Factor: DBP; motif: NRTTAYGTAAYN
|
0.00486373007722665
|
163
|
17
|
3
|
0.176470588235294
|
0.0184049079754601
|
|
TF
|
TF:M00114
|
Factor: Tax/CREB; motif: GGGGGTTGACGYANA
|
0.00488657871929538
|
6548
|
95
|
48
|
0.505263157894737
|
0.00733048259010385
|
|
TF
|
TF:M11271
|
Factor: JunD; motif: NRTGACGCAYN
|
0.00494202949249752
|
7009
|
101
|
53
|
0.524752475247525
|
0.00756170637751462
|
|
TF
|
TF:M11391_1
|
Factor: Erg; motif: NACCGGATATCCGGTN; match class: 1
|
0.00500328037744834
|
9085
|
92
|
59
|
0.641304347826087
|
0.00649422124380848
|
|
TF
|
TF:M07294
|
Factor: KLF8; motif: HMNDGGGTGT
|
0.00502548892072248
|
1361
|
21
|
7
|
0.333333333333333
|
0.00514327700220426
|
|
TF
|
TF:M05318
|
Factor: KLF8; motif: NGGGCGGGGG
|
0.00506690021866325
|
829
|
116
|
14
|
0.120689655172414
|
0.0168878166465621
|
|
TF
|
TF:M05319
|
Factor: KLF7; motif: NGGGCGGGGG
|
0.00506690021866325
|
829
|
116
|
14
|
0.120689655172414
|
0.0168878166465621
|
|
TF
|
TF:M07409
|
Factor: BTEB2; motif: GCCCCRCCCH
|
0.0050888341926475
|
5198
|
96
|
41
|
0.427083333333333
|
0.00788764909580608
|
|
TF
|
TF:M01652_1
|
Factor: p53; motif: RGRCWWGYCYNGRCWWGYYY; match class: 1
|
0.00512560568344085
|
5253
|
101
|
43
|
0.425742574257426
|
0.00818579859128117
|
|
TF
|
TF:M07618
|
Factor: SRF; motif: CCTTWTATGGNN
|
0.00512560568344085
|
167
|
17
|
3
|
0.176470588235294
|
0.0179640718562874
|
|
TF
|
TF:M07356
|
Factor: HIF2A; motif: ACGTGNNN
|
0.00515173907847734
|
1852
|
67
|
16
|
0.238805970149254
|
0.00863930885529158
|
|
TF
|
TF:M01100_1
|
Factor: LRF; motif: GGGGKYNNB; match class: 1
|
0.00521747771690255
|
2762
|
38
|
14
|
0.368421052631579
|
0.00506879073135409
|
|
TF
|
TF:M03922_1
|
Factor: SP4; motif: NWRGCCACGCCCMCTYN; match class: 1
|
0.0053228230660757
|
2143
|
82
|
20
|
0.24390243902439
|
0.00933271115258983
|
|
TF
|
TF:M12151_1
|
Factor: Sp1; motif: NWRGCCMCGCCCMCN; match class: 1
|
0.00534894732653399
|
2548
|
96
|
25
|
0.260416666666667
|
0.0098116169544741
|
|
TF
|
TF:M11273_1
|
Factor: JunB; motif: GATGACGTCAYC; match class: 1
|
0.0053658236766236
|
6582
|
29
|
19
|
0.655172413793103
|
0.00288666058948648
|
|
TF
|
TF:M09734
|
Factor: ZNF692; motif: SYNGGSCCCASCCNC
|
0.0053658236766236
|
11678
|
96
|
73
|
0.760416666666667
|
0.0062510703887652
|
|
TF
|
TF:M12152_1
|
Factor: Sp1; motif: NWRGCCACGCCCMCN; match class: 1
|
0.0053658236766236
|
2859
|
96
|
27
|
0.28125
|
0.00944386149003148
|
|
TF
|
TF:M11313_1
|
Factor: CREBPA; motif: NRTGACGTMANN; match class: 1
|
0.0053658236766236
|
2025
|
34
|
11
|
0.323529411764706
|
0.0054320987654321
|
|
TF
|
TF:M00444_1
|
Factor: VDR; motif: GGGKNARNRRGGWSA; match class: 1
|
0.00537713451475199
|
4534
|
89
|
35
|
0.393258426966292
|
0.00771945302161447
|
|
TF
|
TF:M11218
|
Factor: CREB1; motif: NRTGAYGCGTN
|
0.00538043242865456
|
8461
|
96
|
58
|
0.604166666666667
|
0.00685498168065241
|
|
TF
|
TF:M03920_1
|
Factor: SP1; motif: RCCMCRCCCMC; match class: 1
|
0.00541433289899321
|
3020
|
96
|
28
|
0.291666666666667
|
0.00927152317880795
|
|
TF
|
TF:M00245
|
Factor: Egr-3; motif: NTGCGTGGGCGK
|
0.00549284825057343
|
7313
|
89
|
49
|
0.550561797752809
|
0.00670039655408177
|
|
TF
|
TF:M09668
|
Factor: RelA-p65; motif: GGGRNTTTCCM
|
0.00557165597347027
|
3576
|
63
|
23
|
0.365079365079365
|
0.00643176733780761
|
|
TF
|
TF:M02091
|
Factor: E4F1; motif: RTGACGTAAC
|
0.00557165597347027
|
103
|
120
|
5
|
0.0416666666666667
|
0.0485436893203883
|
|
TF
|
TF:M00041_1
|
Factor: ATF2:c-Jun; motif: TGACGTYA; match class: 1
|
0.00557165597347027
|
1219
|
72
|
13
|
0.180555555555556
|
0.0106644790812141
|
|
TF
|
TF:M11066_1
|
Factor: HES-7; motif: GNCACGYGNN; match class: 1
|
0.00557165597347027
|
8172
|
89
|
53
|
0.595505617977528
|
0.00648556045031816
|
|
TF
|
TF:M09895_1
|
Factor: E2F-6; motif: NGGGCGGGARRNN; match class: 1
|
0.00557165597347027
|
2711
|
81
|
23
|
0.283950617283951
|
0.00848395426042051
|
|
TF
|
TF:M09658_1
|
Factor: Sp2; motif: GGSNNGGGGGCGGGGCCNGNGS; match class: 1
|
0.00562622431738607
|
2033
|
126
|
26
|
0.206349206349206
|
0.0127889818002951
|
|
TF
|
TF:M11228_1
|
Factor: ATF-6; motif: GRTGACGTCAYC; match class: 1
|
0.0056742604556759
|
2102
|
72
|
18
|
0.25
|
0.00856327307326356
|
|
TF
|
TF:M00041
|
Factor: ATF2:c-Jun; motif: TGACGTYA
|
0.00573210831335155
|
4294
|
11
|
8
|
0.727272727272727
|
0.00186306474149977
|
|
TF
|
TF:M11275
|
Factor: XBP-1; motif: SRTGAYGTCAYS
|
0.00573210831335155
|
2714
|
19
|
9
|
0.473684210526316
|
0.00331613854089904
|
|
TF
|
TF:M11395
|
Factor: Erg; motif: NACCGGAWWTCCGGTN
|
0.00581344902062372
|
8715
|
92
|
57
|
0.619565217391304
|
0.00654044750430293
|
|
TF
|
TF:M00649_1
|
Factor: MAZ; motif: GGGGAGGG; match class: 1
|
0.00587305686006478
|
4166
|
79
|
30
|
0.379746835443038
|
0.0072011521843495
|
|
TF
|
TF:M11052_1
|
Factor: Hey2; motif: NNCACGYGNN; match class: 1
|
0.00587305686006478
|
9523
|
89
|
59
|
0.662921348314607
|
0.00619552661976268
|
|
TF
|
TF:M04731_1
|
Factor: Blimp-1; motif: NACTTTCAC; match class: 1
|
0.00587305686006478
|
475
|
7
|
3
|
0.428571428571429
|
0.00631578947368421
|
|
TF
|
TF:M07208_1
|
Factor: EGR1; motif: NCNCCGCCCCCGCN; match class: 1
|
0.00588789082775244
|
2194
|
124
|
27
|
0.217741935483871
|
0.012306289881495
|
|
TF
|
TF:M04826
|
Factor: p300; motif: ACNTCCG
|
0.00594228803243431
|
17806
|
99
|
98
|
0.98989898989899
|
0.00550376277659216
|
|
TF
|
TF:M07312_1
|
Factor: ATF-2; motif: NNTGACGTCAN; match class: 1
|
0.00594228803243431
|
1070
|
72
|
12
|
0.166666666666667
|
0.011214953271028
|
|
TF
|
TF:M10076
|
Factor: SRF; motif: TTNCCWTWTWTGGNCWNN
|
0.00601568361534587
|
187
|
68
|
5
|
0.0735294117647059
|
0.0267379679144385
|
|
TF
|
TF:M00051_1
|
Factor: NF-kappaB; motif: GGGGATYCCC; match class: 1
|
0.00602684972764558
|
2797
|
12
|
7
|
0.583333333333333
|
0.00250268144440472
|
|
TF
|
TF:M12180
|
Factor: BTEB3; motif: NRCCACGCCCMCN
|
0.00602684972764558
|
2088
|
79
|
19
|
0.240506329113924
|
0.00909961685823755
|
|
TF
|
TF:M00444
|
Factor: VDR; motif: GGGKNARNRRGGWSA
|
0.00602684972764558
|
10308
|
95
|
66
|
0.694736842105263
|
0.00640279394644936
|
|
TF
|
TF:M11541_1
|
Factor: Foxn2; motif: NNGCGTCNNNNNGACGCNN; match class: 1
|
0.00612133987180607
|
5493
|
98
|
43
|
0.438775510204082
|
0.00782814491170581
|
|
TF
|
TF:M03976_1
|
Factor: ERG; motif: ACCGGAWATCCGGT; match class: 1
|
0.00614357791673572
|
7781
|
89
|
51
|
0.573033707865168
|
0.00655442745148439
|
|
TF
|
TF:M09817
|
Factor: PAX5; motif: RNGCGTGACCNN
|
0.00614357791673572
|
12511
|
83
|
67
|
0.807228915662651
|
0.00535528734713452
|
|
TF
|
TF:M11535_1
|
Factor: E2F-4; motif: TTTTGGCGCCAWWN; match class: 1
|
0.00614492183929433
|
6532
|
89
|
45
|
0.50561797752809
|
0.00688916105327618
|
|
TF
|
TF:M02279
|
Factor: CREB1; motif: TGACGTCA
|
0.00614492183929433
|
1731
|
34
|
10
|
0.294117647058824
|
0.00577700751010976
|
|
TF
|
TF:M11228
|
Factor: ATF-6; motif: GRTGACGTCAYC
|
0.00614492183929433
|
2123
|
72
|
18
|
0.25
|
0.00847856806406029
|
|
TF
|
TF:M01770
|
Factor: XBP-1; motif: WNNGMCACGTC
|
0.00623009458632275
|
11003
|
98
|
71
|
0.724489795918367
|
0.0064527856039262
|
|
TF
|
TF:M04744_1
|
Factor: ATF-3; motif: GGCGCSSNSNGRTSACGTSA; match class: 1
|
0.00623009458632275
|
4889
|
84
|
35
|
0.416666666666667
|
0.00715892820617713
|
|
TF
|
TF:M11595_1
|
Factor: GCMa; motif: RTGCGGGTN; match class: 1
|
0.00625922784550572
|
5786
|
83
|
39
|
0.469879518072289
|
0.00674040788109229
|
|
TF
|
TF:M11254
|
Factor: AIBZIP; motif: GRTGAYGTCAYC
|
0.00628587649726126
|
1132
|
19
|
6
|
0.315789473684211
|
0.00530035335689046
|
|
TF
|
TF:M12345
|
Factor: Zbtb37; motif: NYACCGCRNTCACCGCR
|
0.00630656229498851
|
5505
|
98
|
43
|
0.438775510204082
|
0.007811080835604
|
|
TF
|
TF:M01692
|
Factor: LTF; motif: GKVACTTNC
|
0.00632872497129972
|
4173
|
69
|
27
|
0.391304347826087
|
0.00647016534867002
|
|
TF
|
TF:M00050
|
Factor: E2F; motif: TTTSGCGC
|
0.00635115342754704
|
7056
|
70
|
39
|
0.557142857142857
|
0.00552721088435374
|
|
TF
|
TF:M11307
|
Factor: ATF-2; motif: NNTGACGTMANN
|
0.00635221018797434
|
2085
|
34
|
11
|
0.323529411764706
|
0.0052757793764988
|
|
TF
|
TF:M12181
|
Factor: BTEB3; motif: NRCCACGCCCMCN
|
0.0063609924950389
|
1597
|
79
|
16
|
0.20253164556962
|
0.0100187852222918
|
|
TF
|
TF:M01177_1
|
Factor: SREBP-2; motif: NNGYCACNNSMN; match class: 1
|
0.00637162604895462
|
2842
|
98
|
27
|
0.275510204081633
|
0.00950035186488388
|
|
TF
|
TF:M11399
|
Factor: Fli-1; motif: NACCGGAWWTCCGGTY
|
0.00642257732496246
|
12977
|
93
|
76
|
0.817204301075269
|
0.00585651537335286
|
|
TF
|
TF:M01588
|
Factor: GKLF; motif: GCCMCRCCCNNN
|
0.00644961483141709
|
8944
|
96
|
60
|
0.625
|
0.00670840787119857
|
|
TF
|
TF:M04950_1
|
Factor: Egr-1; motif: NGCGTGCGY; match class: 1
|
0.00644961483141709
|
5551
|
78
|
36
|
0.461538461538462
|
0.0064853179607278
|
|
TF
|
TF:M08526
|
Factor: E2F-3:HES-7; motif: NNNSGCGCSNNNNNCRCGYGNN
|
0.00651743286218267
|
16867
|
90
|
87
|
0.966666666666667
|
0.00515800083002312
|
|
TF
|
TF:M11266_1
|
Factor: c-Jun; motif: NATGACGTCAYN; match class: 1
|
0.00652094535563346
|
6308
|
22
|
15
|
0.681818181818182
|
0.00237793278376665
|
|
TF
|
TF:M11277
|
Factor: XBP-1; motif: NNTGACGTCAYN
|
0.00652094535563346
|
932
|
72
|
11
|
0.152777777777778
|
0.0118025751072961
|
|
TF
|
TF:M10004
|
Factor: NF-kappaB2; motif: GGGRAANYCCC
|
0.00652604576996107
|
1073
|
63
|
11
|
0.174603174603175
|
0.0102516309412861
|
|
TF
|
TF:M07617_1
|
Factor: Sp4; motif: SCCCCKCCCCCSN; match class: 1
|
0.00654155870737273
|
901
|
124
|
15
|
0.120967741935484
|
0.0166481687014428
|
|
TF
|
TF:M00008_1
|
Factor: Sp1; motif: GGGGCGGGGT; match class: 1
|
0.00674204569822426
|
4937
|
96
|
39
|
0.40625
|
0.00789953413003848
|
|
TF
|
TF:M05439
|
Factor: RREB-1; motif: GGGWCSA
|
0.00678478912452572
|
8635
|
101
|
61
|
0.603960396039604
|
0.00706427330631152
|
|
TF
|
TF:M11695_1
|
Factor: MEF-2D; motif: CCAWATWTRG; match class: 1
|
0.00685084702286425
|
195
|
68
|
5
|
0.0735294117647059
|
0.0256410256410256
|
|
TF
|
TF:M00378_1
|
Factor: Pax-4; motif: NNNNNYCACCCB; match class: 1
|
0.00695556200512659
|
14711
|
96
|
85
|
0.885416666666667
|
0.00577798925973761
|
|
TF
|
TF:M01778
|
Factor: PLAG1; motif: GRGGCNNHNNNRRGGG
|
0.00698697629873114
|
2743
|
44
|
15
|
0.340909090909091
|
0.00546846518410499
|
|
TF
|
TF:M08271
|
Factor: Erm:TBR2; motif: NNGTGNKANNNNNNNNTNNCRSCGGAWGN
|
0.00702774261312045
|
500
|
16
|
4
|
0.25
|
0.008
|
|
TF
|
TF:M05432_1
|
Factor: ZSCAN18; motif: TRRCGRCGNMCC; match class: 1
|
0.00732129235948832
|
818
|
96
|
12
|
0.125
|
0.0146699266503667
|
|
TF
|
TF:M11052
|
Factor: Hey2; motif: NNCACGYGNN
|
0.00741967202637351
|
12502
|
89
|
71
|
0.797752808988764
|
0.00567909134538474
|
|
TF
|
TF:M07329
|
Factor: Osx; motif: CCNCCCCCNNN
|
0.00741967202637351
|
6803
|
89
|
46
|
0.51685393258427
|
0.00676172276936646
|
|
TF
|
TF:M12156
|
Factor: Sp2; motif: NTAAGYCCCGCCCMCTN
|
0.00747627037156046
|
4320
|
88
|
33
|
0.375
|
0.00763888888888889
|
|
TF
|
TF:M07617
|
Factor: Sp4; motif: SCCCCKCCCCCSN
|
0.00759552976169635
|
3595
|
96
|
31
|
0.322916666666667
|
0.0086230876216968
|
|
TF
|
TF:M00215_1
|
Factor: SRF; motif: DCCWTATATGGNCWN; match class: 1
|
0.00766935951417144
|
49
|
68
|
3
|
0.0441176470588235
|
0.0612244897959184
|
|
TF
|
TF:M00738
|
Factor: E2F-4:DP-1; motif: TTTSGCGC
|
0.00768017973719286
|
9812
|
30
|
24
|
0.8
|
0.00244598450876478
|
|
TF
|
TF:M09972
|
Factor: BTEB2; motif: WGGGTGKGGCNGGN
|
0.00782761658283382
|
9217
|
96
|
61
|
0.635416666666667
|
0.0066182054898557
|
|
TF
|
TF:M04950
|
Factor: Egr-1; motif: NGCGTGCGY
|
0.00784909004683502
|
10935
|
85
|
62
|
0.729411764705882
|
0.00566986739826246
|
|
TF
|
TF:M00246
|
Factor: Egr-2; motif: NTGCGTRGGCGK
|
0.00786789006194572
|
6212
|
89
|
43
|
0.48314606741573
|
0.00692208628461043
|
|
TF
|
TF:M11692
|
Factor: MEF-2C; motif: CCAWATWTRG
|
0.0078786606531866
|
1103
|
4
|
3
|
0.75
|
0.00271985494106981
|
|
TF
|
TF:M11533
|
Factor: E2F-1; motif: NTTTTGGCGCCAWWWN
|
0.00789474669908959
|
13105
|
86
|
71
|
0.825581395348837
|
0.0054177794734834
|
|
TF
|
TF:M08952
|
Factor: NF-KAPPAB1; motif: NGGKRNTTYCCCN
|
0.00789474669908959
|
7763
|
21
|
16
|
0.761904761904762
|
0.00206105886899395
|
|
TF
|
TF:M12162
|
Factor: KLF3; motif: GRCCACGCCCN
|
0.00800718718053682
|
1163
|
89
|
14
|
0.157303370786517
|
0.0120378331900258
|
|
TF
|
TF:M10094_1
|
Factor: RelA-p65; motif: NWGGGRATTTCCMN; match class: 1
|
0.00834083862877126
|
242
|
58
|
5
|
0.0862068965517241
|
0.0206611570247934
|
|
TF
|
TF:M10530_1
|
Factor: sp4; motif: NNNGCYCCGCCCCCY; match class: 1
|
0.00834083862877126
|
3780
|
124
|
39
|
0.314516129032258
|
0.0103174603174603
|
|
TF
|
TF:M00514
|
Factor: ATF4; motif: CVTGACGYMABG
|
0.00834083862877126
|
5390
|
84
|
37
|
0.44047619047619
|
0.00686456400742115
|
|
TF
|
TF:M00189_1
|
Factor: AP-2; motif: MKCCCSCNGGCG; match class: 1
|
0.00834463630612229
|
6093
|
96
|
45
|
0.46875
|
0.00738552437223043
|
|
TF
|
TF:M01273
|
Factor: SP4; motif: SCCCCGCCCCS
|
0.00850883365647304
|
3296
|
96
|
29
|
0.302083333333333
|
0.00879854368932039
|
|
TF
|
TF:M03811_1
|
Factor: AP-2gamma; motif: GCCYNCRGSN; match class: 1
|
0.00850883365647304
|
3812
|
84
|
29
|
0.345238095238095
|
0.00760755508919203
|
|
TF
|
TF:M12188_1
|
Factor: KLF17; motif: NGMCMCRCCCTN; match class: 1
|
0.00851368085332939
|
588
|
82
|
9
|
0.109756097560976
|
0.0153061224489796
|
|
TF
|
TF:M01855
|
Factor: AHR; motif: CACGCN
|
0.00871701699041662
|
8491
|
30
|
22
|
0.733333333333333
|
0.00259097868331174
|
|
TF
|
TF:M12148
|
Factor: SP8; motif: NCCACGCCCMCN
|
0.00885011320615142
|
2907
|
79
|
23
|
0.291139240506329
|
0.00791193670450636
|
|
TF
|
TF:M00426
|
Factor: E2F; motif: TTTSGCGS
|
0.00898831476541207
|
11885
|
77
|
60
|
0.779220779220779
|
0.00504838031131679
|
|
TF
|
TF:M11226_1
|
Factor: ATF-6; motif: GRTGACGTCAYC; match class: 1
|
0.00907044425704603
|
825
|
72
|
10
|
0.138888888888889
|
0.0121212121212121
|
|
TF
|
TF:M11223
|
Factor: CREB1; motif: NRTGACGTCANN
|
0.00913999657193993
|
2810
|
72
|
21
|
0.291666666666667
|
0.00747330960854093
|
|
TF
|
TF:M07313_1
|
Factor: ATF-3; motif: SNTGACGYNATN; match class: 1
|
0.00918891204873199
|
544
|
72
|
8
|
0.111111111111111
|
0.0147058823529412
|
|
TF
|
TF:M11257
|
Factor: AIBZIP; motif: NRTGACGTCAYN
|
0.00920337275452949
|
981
|
72
|
11
|
0.152777777777778
|
0.0112130479102956
|
|
TF
|
TF:M10014_1
|
Factor: FXR; motif: NRGGKCANTGRCCNNNNGG; match class: 1
|
0.00948839389047871
|
1498
|
96
|
17
|
0.177083333333333
|
0.0113484646194927
|
|
TF
|
TF:M00193_1
|
Factor: NF-1; motif: NNTTGGCNNNNNNCCNNN; match class: 1
|
0.00948839389047871
|
768
|
20
|
5
|
0.25
|
0.00651041666666667
|
|
TF
|
TF:M00427
|
Factor: E2F; motif: TTTSGCGS
|
0.00952776478103834
|
9485
|
89
|
58
|
0.651685393258427
|
0.00611491829204006
|
|
TF
|
TF:M04926
|
Factor: REST; motif: AAGGTGCT
|
0.00954279088759692
|
6325
|
29
|
18
|
0.620689655172414
|
0.00284584980237154
|
|
TF
|
TF:M03902
|
Factor: GLI2; motif: GCGACCACMCTR
|
0.00954440924021914
|
1567
|
61
|
13
|
0.213114754098361
|
0.00829610721123165
|
|
TF
|
TF:M12153_1
|
Factor: Sp3; motif: NGCCACGCCCMCN; match class: 1
|
0.00960842034005892
|
1938
|
82
|
18
|
0.219512195121951
|
0.00928792569659443
|
|
TF
|
TF:M00193
|
Factor: NF-1; motif: NNTTGGCNNNNNNCCNNN
|
0.00960842034005892
|
5034
|
66
|
29
|
0.439393939393939
|
0.00576082638061184
|
|
TF
|
TF:M08980
|
Factor: VDR:RXR-ALPHA; motif: NRGGTCANNNGGTTCNN
|
0.00960842034005892
|
10442
|
19
|
17
|
0.894736842105263
|
0.00162804060524804
|
|
TF
|
TF:M02108
|
Factor: CSX; motif: NNCACTTGNRN
|
0.00968974990850611
|
7061
|
19
|
14
|
0.736842105263158
|
0.00198272199405183
|
|
TF
|
TF:M12149
|
Factor: SP9; motif: RCCACGCCCMCY
|
0.00989652306357265
|
2379
|
79
|
20
|
0.253164556962025
|
0.00840689365279529
|
|
TF
|
TF:M11231_1
|
Factor: CREBL1; motif: GRTGACGTCAYC; match class: 1
|
0.0102028431583902
|
841
|
72
|
10
|
0.138888888888889
|
0.0118906064209275
|
|
TF
|
TF:M11223_1
|
Factor: CREB1; motif: NRTGACGTCANN; match class: 1
|
0.0102670378802435
|
2678
|
29
|
11
|
0.379310344827586
|
0.0041075429424944
|
|
TF
|
TF:M04710_1
|
Factor: CHD2; motif: TCTCGCGAG; match class: 1
|
0.0103749629917983
|
11477
|
96
|
71
|
0.739583333333333
|
0.00618628561470768
|
|
TF
|
TF:M11877
|
Factor: pax-2; motif: NCGTCACGCNYSRNYGCNYN
|
0.0105006066835652
|
11523
|
26
|
23
|
0.884615384615385
|
0.00199600798403194
|
|
TF
|
TF:M11226
|
Factor: ATF-6; motif: GRTGACGTCAYC
|
0.010502668218021
|
845
|
72
|
10
|
0.138888888888889
|
0.0118343195266272
|
|
TF
|
TF:M12354
|
Factor: ZNF37A; motif: CCYYGGCTCCNTSCCMN
|
0.0105378364860054
|
10961
|
84
|
61
|
0.726190476190476
|
0.00556518565824286
|
|
TF
|
TF:M09817_1
|
Factor: PAX5; motif: RNGCGTGACCNN; match class: 1
|
0.0106187574389872
|
6170
|
96
|
45
|
0.46875
|
0.00729335494327391
|
|
TF
|
TF:M04193
|
Factor: TFE3; motif: NNCACGTGAY
|
0.0106976295627779
|
2273
|
83
|
20
|
0.240963855421687
|
0.0087989441267048
|
|
TF
|
TF:M12119_1
|
Factor: FPM315; motif: NGTGCTCCCN; match class: 1
|
0.0107076421170525
|
61
|
63
|
3
|
0.0476190476190476
|
0.0491803278688525
|
|
TF
|
TF:M07329_1
|
Factor: Osx; motif: CCNCCCCCNNN; match class: 1
|
0.0107387873127926
|
2509
|
126
|
29
|
0.23015873015873
|
0.0115583897967318
|
|
TF
|
TF:M04146
|
Factor: TFAP2A; motif: YGCCCNNRGGCN
|
0.0107441293184193
|
6552
|
96
|
47
|
0.489583333333333
|
0.00717338217338217
|
|
TF
|
TF:M11656_1
|
Factor: HSF1; motif: RGAANRTTCYRGAAN; match class: 1
|
0.0107441293184193
|
219
|
40
|
4
|
0.1
|
0.0182648401826484
|
|
TF
|
TF:M12163
|
Factor: KLF3; motif: GRCCRCGCCCN
|
0.0107813388952093
|
733
|
69
|
9
|
0.130434782608696
|
0.0122783083219645
|
|
TF
|
TF:M00806_1
|
Factor: NF-1; motif: NTGGNNNNNNGCCAANN; match class: 1
|
0.010872522923254
|
562
|
41
|
6
|
0.146341463414634
|
0.0106761565836299
|
|
TF
|
TF:M02279_1
|
Factor: CREB1; motif: TGACGTCA; match class: 1
|
0.0108807540814662
|
851
|
72
|
10
|
0.138888888888889
|
0.0117508813160987
|
|
TF
|
TF:M01862_1
|
Factor: ATF-2; motif: NNNTGACGTNAN; match class: 1
|
0.0108807540814662
|
1007
|
72
|
11
|
0.152777777777778
|
0.0109235352532274
|
|
TF
|
TF:M01047_1
|
Factor: AP-2alphaA; motif: ANMGCCTNAGGCKNT; match class: 1
|
0.0108871555639253
|
9237
|
86
|
55
|
0.63953488372093
|
0.00595431417126773
|
|
TF
|
TF:M07249
|
Factor: ctcf; motif: CCNCNAGRKGGCRSTN
|
0.0108871555639253
|
6715
|
75
|
39
|
0.52
|
0.00580789277736411
|
|
TF
|
TF:M01047
|
Factor: AP-2alphaA; motif: ANMGCCTNAGGCKNT
|
0.0108871555639253
|
9237
|
86
|
55
|
0.63953488372093
|
0.00595431417126773
|
|
TF
|
TF:M09765_1
|
Factor: SP1; motif: NRGKGGGCGGGGCN; match class: 1
|
0.0108871555639253
|
3318
|
124
|
35
|
0.282258064516129
|
0.0105485232067511
|
|
TF
|
TF:M09638
|
Factor: MEF-2C; motif: NKCTATTTTTRGN
|
0.0112334401141124
|
1106
|
47
|
9
|
0.191489361702128
|
0.0081374321880651
|
|
TF
|
TF:M08523
|
Factor: E2F-1:TBR2; motif: NGGTGNNANGGCGCNNTNNCRNNN
|
0.0112395397053366
|
12160
|
89
|
69
|
0.775280898876405
|
0.00567434210526316
|
|
TF
|
TF:M11498_1
|
Factor: OC-2; motif: NNATCGATYN; match class: 1
|
0.0112880863364952
|
396
|
58
|
6
|
0.103448275862069
|
0.0151515151515152
|
|
TF
|
TF:M03971
|
Factor: ELK1; motif: NACTTCCGSCGGAAGYN
|
0.0113680216074007
|
11293
|
96
|
70
|
0.729166666666667
|
0.0061985300628708
|
|
TF
|
TF:M09893_1
|
Factor: E2F-3; motif: NNGGCGGGAAA; match class: 1
|
0.0114814775360208
|
1322
|
101
|
16
|
0.158415841584158
|
0.0121028744326778
|
|
TF
|
TF:M09632
|
Factor: Kaiso; motif: GNTCTCGCGAGRNNNNGGN
|
0.0117653931489062
|
3001
|
88
|
25
|
0.284090909090909
|
0.00833055648117294
|
|
TF
|
TF:M12085
|
Factor: ZNF501; motif: NNNCSACGCGAACAC
|
0.0117653931489062
|
8467
|
45
|
30
|
0.666666666666667
|
0.00354316759182709
|
|
TF
|
TF:M11298_1
|
Factor: jdp2; motif: NRTGACGTCAYN; match class: 1
|
0.0119002773439391
|
864
|
72
|
10
|
0.138888888888889
|
0.0115740740740741
|
|
TF
|
TF:M08440
|
Factor: AP-2gamma:E2F-8; motif: NTTCCCGCNNSCCCSMGGC
|
0.0120656444540783
|
1200
|
122
|
17
|
0.139344262295082
|
0.0141666666666667
|
|
TF
|
TF:M11285
|
Factor: Fra-1; motif: GRTGACGTCATC
|
0.0121602406735097
|
7708
|
31
|
21
|
0.67741935483871
|
0.0027244421380384
|
|
TF
|
TF:M01298
|
Factor: MECP2; motif: CCGGSNNNANNAWWT
|
0.0124166949002128
|
1290
|
58
|
11
|
0.189655172413793
|
0.00852713178294574
|
|
TF
|
TF:M08660
|
Factor: AP-2gamma:Dlx-3; motif: NSCCYNNRGGCANNNNYAATTA
|
0.0124166949002128
|
3566
|
100
|
31
|
0.31
|
0.0086932136848009
|
|
TF
|
TF:M04109_1
|
Factor: RUNX3; motif: NRACCGCAAACCGCAN; match class: 1
|
0.0124869597284532
|
3878
|
124
|
39
|
0.314516129032258
|
0.0100567302733368
|
|
TF
|
TF:M11304
|
Factor: batf3; motif: NRTGAYGTCAYN
|
0.0125795789870725
|
763
|
43
|
7
|
0.162790697674419
|
0.00917431192660551
|
|
TF
|
TF:M04748_1
|
Factor: GABP-alpha; motif: AACCGGAAR; match class: 1
|
0.0126376965168134
|
10368
|
100
|
68
|
0.68
|
0.00655864197530864
|
|
TF
|
TF:M03544_1
|
Factor: CREB1; motif: NNNNACGTCANN; match class: 1
|
0.012703076323972
|
1196
|
72
|
12
|
0.166666666666667
|
0.0100334448160535
|
|
TF
|
TF:M09968
|
Factor: KLF15; motif: RGGGMGGRGNNGGGGGNGG
|
0.0128224683026618
|
3157
|
80
|
24
|
0.3
|
0.00760215394361736
|
|
TF
|
TF:M09640_1
|
Factor: NF-1C; motif: NYYTGGCWNNNNKCCMN; match class: 1
|
0.0130166886446985
|
337
|
123
|
8
|
0.0650406504065041
|
0.0237388724035608
|
|
TF
|
TF:M11303_1
|
Factor: batf3; motif: GRTGACGTCATC; match class: 1
|
0.013184685544947
|
584
|
72
|
8
|
0.111111111111111
|
0.0136986301369863
|
|
TF
|
TF:M01175_1
|
Factor: CKROX; motif: SCCCTCCCC; match class: 1
|
0.0133308680194507
|
2679
|
83
|
22
|
0.265060240963855
|
0.0082120194102277
|
|
TF
|
TF:M04196
|
Factor: USF1; motif: RNCACGTGAY
|
0.0133621228394938
|
1816
|
83
|
17
|
0.204819277108434
|
0.00936123348017621
|
|
TF
|
TF:M04748
|
Factor: GABP-alpha; motif: AACCGGAAR
|
0.0133716223875598
|
16027
|
32
|
32
|
1
|
0.00199663068571785
|
|
TF
|
TF:M07350
|
Factor: BMAL1; motif: AGCCAYKKGA
|
0.0137573116390991
|
1392
|
89
|
15
|
0.168539325842697
|
0.0107758620689655
|
|
TF
|
TF:M12354_1
|
Factor: ZNF37A; motif: CCYYGGCTCCNTSCCMN; match class: 1
|
0.0137573116390991
|
5624
|
100
|
43
|
0.43
|
0.00764580369843528
|
|
TF
|
TF:M03545_1
|
Factor: c-Rel; motif: NGGGAATYTCCN; match class: 1
|
0.013866150747063
|
1011
|
43
|
8
|
0.186046511627907
|
0.00791295746785361
|
|
TF
|
TF:M11277_1
|
Factor: XBP-1; motif: NNTGACGTCAYN; match class: 1
|
0.0139241144819717
|
886
|
72
|
10
|
0.138888888888889
|
0.0112866817155756
|
|
TF
|
TF:M12147
|
Factor: SP8; motif: RNCACGCCCMCN
|
0.0141424839265164
|
2465
|
79
|
20
|
0.253164556962025
|
0.00811359026369168
|
|
TF
|
TF:M07248_1
|
Factor: CREB1; motif: NNNNTGACGTNANNN; match class: 1
|
0.0143280628733327
|
1216
|
72
|
12
|
0.166666666666667
|
0.00986842105263158
|
|
TF
|
TF:M08208
|
Factor: E2F-3:FOXI1; motif: NGACACCGCGCCCAC
|
0.0143687456072775
|
12560
|
38
|
33
|
0.868421052631579
|
0.00262738853503185
|
|
TF
|
TF:M07313
|
Factor: ATF-3; motif: SNTGACGYNATN
|
0.0144996145778698
|
2894
|
73
|
21
|
0.287671232876712
|
0.00725639253628196
|
|
TF
|
TF:M09987
|
Factor: MEF-2D; motif: NKCTATTTTTAG
|
0.0147696365686477
|
427
|
11
|
3
|
0.272727272727273
|
0.00702576112412178
|
|
TF
|
TF:M07358
|
Factor: HSF2; motif: NGGAATNTTCTNGNRN
|
0.0148014462716116
|
576
|
92
|
9
|
0.0978260869565217
|
0.015625
|
|
TF
|
TF:M09764
|
Factor: ROX; motif: GNNNCASGTGGS
|
0.0148183864302261
|
5809
|
92
|
41
|
0.445652173913043
|
0.00705801342744018
|
|
TF
|
TF:M10151
|
Factor: EZI; motif: GGGGAGGGGRRGGRGRRGGRRR
|
0.0149586902787871
|
2108
|
92
|
20
|
0.217391304347826
|
0.0094876660341556
|
|
TF
|
TF:M11231
|
Factor: CREBL1; motif: GRTGACGTCAYC
|
0.0149586902787871
|
1057
|
72
|
11
|
0.152777777777778
|
0.010406811731315
|
|
TF
|
TF:M11308_1
|
Factor: ATF-2; motif: NRTGAYGTMAYN; match class: 1
|
0.0150287448626433
|
1225
|
72
|
12
|
0.166666666666667
|
0.00979591836734694
|
|
TF
|
TF:M08905_1
|
Factor: TIEG1; motif: GGSGGKGNNN; match class: 1
|
0.0150287448626433
|
1017
|
34
|
7
|
0.205882352941176
|
0.00688298918387414
|
|
TF
|
TF:M00341
|
Factor: GABP; motif: VCCGGAAGNGCR
|
0.0151378695973158
|
8224
|
86
|
50
|
0.581395348837209
|
0.00607976653696498
|
|
TF
|
TF:M11257_1
|
Factor: AIBZIP; motif: NRTGACGTCAYN; match class: 1
|
0.0151722281106526
|
899
|
72
|
10
|
0.138888888888889
|
0.0111234705228031
|
|
TF
|
TF:M11215_1
|
Factor: N-Myc; motif: NNCACGTGNN; match class: 1
|
0.0152878016094915
|
67
|
67
|
3
|
0.0447761194029851
|
0.0447761194029851
|
|
TF
|
TF:M07384_1
|
Factor: HIF-1alpha; motif: NCACGTNN; match class: 1
|
0.0152878016094915
|
67
|
67
|
3
|
0.0447761194029851
|
0.0447761194029851
|
|
TF
|
TF:M10115_1
|
Factor: LRF; motif: NRGGGKCKY; match class: 1
|
0.0153997274628644
|
2326
|
96
|
22
|
0.229166666666667
|
0.00945829750644884
|
|
TF
|
TF:M00272
|
Factor: p53; motif: NGRCWTGYCY
|
0.0154491614605134
|
4151
|
24
|
12
|
0.5
|
0.00289086966995905
|
|
TF
|
TF:M11221
|
Factor: CREB1; motif: RTGACGYGTCAN
|
0.0154510137261503
|
9677
|
89
|
58
|
0.651685393258427
|
0.00599359305569908
|
|
TF
|
TF:M08725
|
Factor: meis1:Max; motif: NTGACRNNNNNNCACGTGC
|
0.015481365464605
|
906
|
84
|
11
|
0.130952380952381
|
0.0121412803532009
|
|
TF
|
TF:M03554
|
Factor: NF-1A; motif: NGCCARN
|
0.0155556303578792
|
4908
|
16
|
10
|
0.625
|
0.00203748981255094
|
|
TF
|
TF:M12232
|
Factor: c-Krox; motif: NCGACCACCN
|
0.0157231679183382
|
2982
|
81
|
23
|
0.283950617283951
|
0.00771294433266264
|
|
TF
|
TF:M00039_1
|
Factor: CREB; motif: TGACGTMA; match class: 1
|
0.0157265974123463
|
1482
|
4
|
3
|
0.75
|
0.00202429149797571
|
|
TF
|
TF:M00797
|
Factor: HIF1; motif: GNNKACGTGCGGNN
|
0.0159856780574265
|
3576
|
89
|
28
|
0.314606741573034
|
0.00782997762863535
|
|
TF
|
TF:M09668_1
|
Factor: RelA-p65; motif: GGGRNTTTCCM; match class: 1
|
0.0159856780574265
|
513
|
49
|
6
|
0.122448979591837
|
0.0116959064327485
|
|
TF
|
TF:M01865_1
|
Factor: BTEB3; motif: BNRNGGGAGGNGT; match class: 1
|
0.0159856780574265
|
4218
|
49
|
20
|
0.408163265306122
|
0.00474158368895211
|
|
TF
|
TF:M00646_1
|
Factor: LF-A1; motif: GGGSTCWR; match class: 1
|
0.016013741327345
|
6320
|
37
|
21
|
0.567567567567568
|
0.00332278481012658
|
|
TF
|
TF:M00054_1
|
Factor: NF-kappaB; motif: GGGAMTTYCC; match class: 1
|
0.0161780503876698
|
712
|
123
|
12
|
0.0975609756097561
|
0.0168539325842697
|
|
TF
|
TF:M08993_1
|
Factor: ZNF777; motif: GTCCGYCCCGTCSAACAAT; match class: 1
|
0.0161780503876698
|
4112
|
82
|
29
|
0.353658536585366
|
0.00705252918287938
|
|
TF
|
TF:M07277_1
|
Factor: BTEB2; motif: RGGGNGKGGN; match class: 1
|
0.0163578518246152
|
4149
|
96
|
33
|
0.34375
|
0.00795372378886479
|
|
TF
|
TF:M08941_1
|
Factor: C-JUN:FRA-2; motif: NRTGACGTMAT; match class: 1
|
0.0164928426803187
|
3353
|
29
|
12
|
0.413793103448276
|
0.00357888458097226
|
|
TF
|
TF:M00104_1
|
Factor: CDP; motif: NATCGATCGS; match class: 1
|
0.0168215192382888
|
1353
|
58
|
11
|
0.189655172413793
|
0.00813008130081301
|
|
TF
|
TF:M11303
|
Factor: batf3; motif: GRTGACGTCATC
|
0.0168872831420627
|
614
|
72
|
8
|
0.111111111111111
|
0.0130293159609121
|
|
TF
|
TF:M03574
|
Factor: GCMa; motif: CCCGCAT
|
0.0169319625549612
|
2362
|
61
|
16
|
0.262295081967213
|
0.00677392040643522
|
|
TF
|
TF:M11417_1
|
Factor: c-Ets-2; motif: NACCGGAAGYRCTTCCGGTN; match class: 1
|
0.0170227687619422
|
14041
|
92
|
78
|
0.847826086956522
|
0.0055551598888968
|
|
TF
|
TF:M03978
|
Factor: ETS1; motif: GCCGGAWGTACTTCCGGN
|
0.0171827485238861
|
12082
|
89
|
68
|
0.764044943820225
|
0.00562820725045522
|
|
TF
|
TF:M04016
|
Factor: IRF5; motif: CCGAAACCGAAACY
|
0.0171917685702459
|
314
|
33
|
4
|
0.121212121212121
|
0.0127388535031847
|
|
TF
|
TF:M04797_1
|
Factor: Egr-1; motif: CCGCCCMCG; match class: 1
|
0.0175020493812003
|
1565
|
82
|
15
|
0.182926829268293
|
0.00958466453674121
|
|
TF
|
TF:M11275_1
|
Factor: XBP-1; motif: SRTGAYGTCAYS; match class: 1
|
0.0176718121222761
|
923
|
72
|
10
|
0.138888888888889
|
0.0108342361863489
|
|
TF
|
TF:M11251_1
|
Factor: Luman; motif: NRTGACGTCAYN; match class: 1
|
0.0177583141056566
|
768
|
72
|
9
|
0.125
|
0.01171875
|
|
TF
|
TF:M04013
|
Factor: HSFY1; motif: TTCGAANSRTTCGAA
|
0.0177583141056566
|
962
|
58
|
9
|
0.155172413793103
|
0.00935550935550936
|
|
TF
|
TF:M01714
|
Factor: KLF15; motif: GAGNNGGGGNGTDG
|
0.0178647707417747
|
2159
|
98
|
21
|
0.214285714285714
|
0.00972672533580361
|
|
TF
|
TF:M11308
|
Factor: ATF-2; motif: NRTGAYGTMAYN
|
0.0179773709811241
|
4263
|
21
|
11
|
0.523809523809524
|
0.00258034248182031
|
|
TF
|
TF:M07063_1
|
Factor: Sp1; motif: GGGGCGGGGC; match class: 1
|
0.0179773709811241
|
3495
|
96
|
29
|
0.302083333333333
|
0.00829756795422031
|
|
TF
|
TF:M10435_1
|
Factor: Sp2; motif: GGGGCGGGG; match class: 1
|
0.0179773709811241
|
3495
|
96
|
29
|
0.302083333333333
|
0.00829756795422031
|
|
TF
|
TF:M09985
|
Factor: Mef-2a; motif: NKCTATTTTTRGN
|
0.0181658477226407
|
1454
|
47
|
10
|
0.212765957446809
|
0.00687757909215956
|
|
TF
|
TF:M01858
|
Factor: AP-2beta; motif: GCNNNGGSCNGVGGGN
|
0.0182432349259658
|
7418
|
86
|
46
|
0.534883720930233
|
0.00620113238069561
|
|
TF
|
TF:M08930
|
Factor: JUND:FRA-1; motif: RTGACGTMAY
|
0.0182432349259658
|
5702
|
59
|
28
|
0.474576271186441
|
0.00491055769905296
|
|
TF
|
TF:M12150
|
Factor: SP9; motif: NCCACGCCCMCN
|
0.0187155811320995
|
2353
|
79
|
19
|
0.240506329113924
|
0.00807479813004675
|
|
TF
|
TF:M07348_1
|
Factor: AP-2alpha; motif: NSCCNCRGGSN; match class: 1
|
0.0187823689993389
|
2945
|
98
|
26
|
0.26530612244898
|
0.00882852292020373
|
|
TF
|
TF:M11250
|
Factor: Luman; motif: NGCCACGTCAYN
|
0.0188548830647477
|
223
|
79
|
5
|
0.0632911392405063
|
0.0224215246636771
|
|
TF
|
TF:M11225_1
|
Factor: CREM; motif: NNTGACGTCANN; match class: 1
|
0.0188548830647477
|
2482
|
29
|
10
|
0.344827586206897
|
0.0040290088638195
|
|
TF
|
TF:M00938_1
|
Factor: E2F-1; motif: TTGGCGCGRAANNGNM; match class: 1
|
0.0188548830647477
|
2573
|
89
|
22
|
0.247191011235955
|
0.00855033035367276
|
|
TF
|
TF:M04110_1
|
Factor: RUNX3; motif: NRACCGCANWAACCRCAN; match class: 1
|
0.018858649819144
|
3764
|
86
|
28
|
0.325581395348837
|
0.00743889479277365
|
|
TF
|
TF:M07129_1
|
Factor: Sp2; motif: GYCCCGCCYCYNNNN; match class: 1
|
0.018858649819144
|
2422
|
124
|
27
|
0.217741935483871
|
0.0111478117258464
|
|
TF
|
TF:M00665
|
Factor: Sp3; motif: ASMCTTGGGSRGGG
|
0.0189227121225728
|
7729
|
81
|
45
|
0.555555555555556
|
0.00582222797257084
|
|
TF
|
TF:M04691_1
|
Factor: Kaiso; motif: TCTCGCGAG; match class: 1
|
0.0190374819080094
|
10488
|
36
|
28
|
0.777777777777778
|
0.0026697177726926
|
|
TF
|
TF:M08869
|
Factor: CLOCKBMAL; motif: NNNWCACGTGGN
|
0.0190374819080094
|
1848
|
85
|
17
|
0.2
|
0.0091991341991342
|
|
TF
|
TF:M11251
|
Factor: Luman; motif: NRTGACGTCAYN
|
0.0190994528815956
|
780
|
72
|
9
|
0.125
|
0.0115384615384615
|
|
TF
|
TF:M00257_1
|
Factor: RREB-1; motif: CCCCAAACMMCCCC; match class: 1
|
0.0190994528815956
|
4475
|
94
|
34
|
0.361702127659574
|
0.00759776536312849
|
|
TF
|
TF:M00236
|
Factor: Arnt; motif: NDDNNCACGTGNNNNN
|
0.0191994167952954
|
2210
|
60
|
15
|
0.25
|
0.00678733031674208
|
|
TF
|
TF:M09986
|
Factor: MEF-2C; motif: NKCTATTTTTRGN
|
0.0193560412065558
|
1072
|
11
|
4
|
0.363636363636364
|
0.00373134328358209
|
|
TF
|
TF:M08874
|
Factor: E2F1; motif: NNNNNGCGSSAAAN
|
0.0195837411119946
|
7201
|
75
|
40
|
0.533333333333333
|
0.00555478405776975
|
|
TF
|
TF:M08980_1
|
Factor: VDR:RXR-ALPHA; motif: NRGGTCANNNGGTTCNN; match class: 1
|
0.0198611042789142
|
3939
|
79
|
27
|
0.341772151898734
|
0.00685453160700685
|
|
TF
|
TF:M02012_1
|
Factor: HIF-1alpha; motif: NCACGT; match class: 1
|
0.0199214257607064
|
1770
|
67
|
14
|
0.208955223880597
|
0.00790960451977401
|
|
TF
|
TF:M12219
|
Factor: ZIC1; motif: NACCCCCCGCTGYGC
|
0.0199214257607064
|
832
|
95
|
11
|
0.115789473684211
|
0.0132211538461538
|
|
TF
|
TF:M00427_1
|
Factor: E2F; motif: TTTSGCGS; match class: 1
|
0.0199891311140648
|
3831
|
85
|
28
|
0.329411764705882
|
0.00730879665883581
|
|
TF
|
TF:M11559
|
Factor: FKHL14; motif: NYAMAYAAACAN
|
0.0200023242667838
|
2701
|
70
|
19
|
0.271428571428571
|
0.00703443169196594
|
|
TF
|
TF:M00513_1
|
Factor: ATF3; motif: CBCTGACGTCANCS; match class: 1
|
0.0200414855373886
|
3374
|
19
|
9
|
0.473684210526316
|
0.0026674570243035
|
|
TF
|
TF:M11306
|
Factor: ATF-2; motif: NRTGAYGTMAYN
|
0.0201842279305056
|
3378
|
19
|
9
|
0.473684210526316
|
0.00266429840142096
|
|
TF
|
TF:M11224
|
Factor: CREM; motif: NRTGATGTCAYN
|
0.0203277659540149
|
3382
|
19
|
9
|
0.473684210526316
|
0.00266114725014784
|
|
TF
|
TF:M11655
|
Factor: HSF1; motif: NGAANGTTCKRGAAN
|
0.020690305906034
|
1207
|
97
|
14
|
0.144329896907216
|
0.0115990057995029
|
|
TF
|
TF:M12135_1
|
Factor: mbnl2; motif: NYGCYTYGCTTN; match class: 1
|
0.0207349507171241
|
1097
|
11
|
4
|
0.363636363636364
|
0.00364630811303555
|
|
TF
|
TF:M08224
|
Factor: Elk-1:Pax-1; motif: ACCGGAACTACGCWTSANTG
|
0.0210198526988329
|
8925
|
11
|
10
|
0.909090909090909
|
0.00112044817927171
|
|
TF
|
TF:M09877
|
Factor: BMAL1; motif: NSCACGTGNSN
|
0.0210996701647046
|
2189
|
92
|
20
|
0.217391304347826
|
0.00913659205116491
|
|
TF
|
TF:M03914_1
|
Factor: KLF16; motif: GMCACGCCCCC; match class: 1
|
0.0214877909491177
|
710
|
96
|
10
|
0.104166666666667
|
0.0140845070422535
|
|
TF
|
TF:M09890
|
Factor: ctcf; motif: YGGCCACCAGRKGGCRSYN
|
0.021567501930462
|
8479
|
101
|
58
|
0.574257425742574
|
0.00684042929590754
|
|
TF
|
TF:M08867
|
Factor: AP2; motif: GCCYGSGGSN
|
0.0217859305477298
|
10054
|
101
|
66
|
0.653465346534653
|
0.00656455142231947
|
|
TF
|
TF:M11654
|
Factor: HSF1; motif: RGAANRTTCYRGAAN
|
0.0219033437651251
|
1353
|
97
|
15
|
0.154639175257732
|
0.0110864745011086
|
|
TF
|
TF:M00232
|
Factor: MEF-2A; motif: NNNNNWKCTAWAAATAGMNNNN
|
0.0219033437651251
|
1421
|
27
|
7
|
0.259259259259259
|
0.00492610837438424
|
|
TF
|
TF:M11568
|
Factor: HNF-3alpha; motif: NNYTAWGTAAACAAAN
|
0.0219916965504795
|
2277
|
89
|
20
|
0.224719101123595
|
0.00878348704435661
|
|
TF
|
TF:M10158_1
|
Factor: ZNF563; motif: NGNNTCMTNNCNGGCAGCTGY; match class: 1
|
0.022452264112013
|
1310
|
81
|
13
|
0.160493827160494
|
0.0099236641221374
|
|
TF
|
TF:M08226_1
|
Factor: Elk-1:Pax-9; motif: ACCGGAACYACGCWYSANTG; match class: 1
|
0.0225345004630785
|
5604
|
22
|
13
|
0.590909090909091
|
0.0023197715917202
|
|
TF
|
TF:M11296_1
|
Factor: jdp2; motif: NRTGAYGTCAYN; match class: 1
|
0.0230187159673935
|
967
|
72
|
10
|
0.138888888888889
|
0.0103412616339193
|
|
TF
|
TF:M00084_1
|
Factor: MZF-1; motif: KNGNKAGGGGNAA; match class: 1
|
0.0230408704873497
|
1683
|
87
|
16
|
0.183908045977011
|
0.0095068330362448
|
|
TF
|
TF:M12179
|
Factor: BTEB3; motif: NRCCACGCCCMCN
|
0.0230408704873497
|
1350
|
79
|
13
|
0.164556962025316
|
0.00962962962962963
|
|
TF
|
TF:M07297
|
Factor: MAZ; motif: CCCTCCCYCYN
|
0.0231034412915972
|
2964
|
38
|
13
|
0.342105263157895
|
0.0043859649122807
|
|
TF
|
TF:M04212
|
Factor: HLF; motif: NNTTACRTAAYN
|
0.023379787083415
|
330
|
17
|
3
|
0.176470588235294
|
0.00909090909090909
|
|
TF
|
TF:M00052
|
Factor: NF-kappaB; motif: GGGRATTTCC
|
0.0244332869778303
|
1874
|
72
|
15
|
0.208333333333333
|
0.0080042689434365
|
|
TF
|
TF:M01154_1
|
Factor: c-Myc; motif: KACCACGTGSYY; match class: 1
|
0.024447303801885
|
3751
|
92
|
29
|
0.315217391304348
|
0.00773127166089043
|
|
TF
|
TF:M10530
|
Factor: sp4; motif: NNNGCYCCGCCCCCY
|
0.024554007531667
|
8227
|
96
|
54
|
0.5625
|
0.00656375349459098
|
|
TF
|
TF:M11081_1
|
Factor: SREBP-1; motif: RTCRCGTGAY; match class: 1
|
0.0247386355604595
|
7890
|
89
|
49
|
0.550561797752809
|
0.00621039290240811
|
|
TF
|
TF:M01858_1
|
Factor: AP-2beta; motif: GCNNNGGSCNGVGGGN; match class: 1
|
0.0247386355604595
|
3141
|
85
|
24
|
0.282352941176471
|
0.00764087870105062
|
|
TF
|
TF:M04710
|
Factor: CHD2; motif: TCTCGCGAG
|
0.0247386355604595
|
14109
|
86
|
73
|
0.848837209302326
|
0.00517400240980934
|
|
TF
|
TF:M00740
|
Factor: Rb:E2F-1:DP-1; motif: TTTSGCGC
|
0.0248454652586669
|
10427
|
95
|
64
|
0.673684210526316
|
0.00613791119209744
|
|
TF
|
TF:M11221_1
|
Factor: CREB1; motif: RTGACGYGTCAN; match class: 1
|
0.0250782506667206
|
5695
|
88
|
38
|
0.431818181818182
|
0.00667251975417033
|
|
TF
|
TF:M11289
|
Factor: ATF-3; motif: NRTGACGTCAYN
|
0.0252755760345815
|
4773
|
31
|
15
|
0.483870967741935
|
0.00314267756128221
|
|
TF
|
TF:M09867
|
Factor: ATF-2; motif: RRTGANGTCAY
|
0.0252755760345815
|
4328
|
16
|
9
|
0.5625
|
0.00207948243992606
|
|
TF
|
TF:M12172_1
|
Factor: LKLF; motif: NRCCACRCCCN; match class: 1
|
0.0252755760345815
|
243
|
79
|
5
|
0.0632911392405063
|
0.0205761316872428
|
|
TF
|
TF:M04934
|
Factor: TR4; motif: ACCCCGS
|
0.0252755760345815
|
18103
|
68
|
68
|
1
|
0.00375628348892449
|
|
TF
|
TF:M12230_1
|
Factor: LRF; motif: RCGACCACCNN; match class: 1
|
0.0252755760345815
|
2637
|
68
|
18
|
0.264705882352941
|
0.0068259385665529
|
|
TF
|
TF:M09602
|
Factor: ctcf; motif: NNNCCASYAGRKGGCRSYNN
|
0.0254356893858199
|
7450
|
96
|
50
|
0.520833333333333
|
0.00671140939597315
|
|
TF
|
TF:M11018
|
Factor: IRX-1; motif: NACRYNNNNNNNNRYGNN
|
0.0255545235731767
|
16955
|
90
|
86
|
0.955555555555556
|
0.00507225007372457
|
|
TF
|
TF:M07043
|
Factor: HIF-1alpha; motif: NNACGTGNN
|
0.0262621561276274
|
1439
|
67
|
12
|
0.17910447761194
|
0.00833912439193885
|
|
TF
|
TF:M11508_1
|
Factor: CDP; motif: NNATYGATYN; match class: 1
|
0.0265580390163299
|
1591
|
61
|
12
|
0.19672131147541
|
0.00754242614707731
|
|
TF
|
TF:M05386
|
Factor: KLF17; motif: NGGGCGG
|
0.0268647762853865
|
7900
|
85
|
47
|
0.552941176470588
|
0.00594936708860759
|
|
TF
|
TF:M05499
|
Factor: LKLF; motif: NGGGCGG
|
0.0268647762853865
|
7900
|
85
|
47
|
0.552941176470588
|
0.00594936708860759
|
|
TF
|
TF:M08266
|
Factor: ER71:Pax-5; motif: ACCGGAACYACGCWTSANTG
|
0.0272431072751643
|
14377
|
90
|
77
|
0.855555555555556
|
0.0053557765876052
|
|
TF
|
TF:M07615_1
|
Factor: Sp3; motif: GGGGCGGGGSNN; match class: 1
|
0.0272891494039245
|
2885
|
124
|
30
|
0.241935483870968
|
0.0103986135181976
|
|
TF
|
TF:M00237
|
Factor: AhR:Arnt; motif: GRGKATYGCGTGMCWNSCC
|
0.0279489684762309
|
6327
|
96
|
44
|
0.458333333333333
|
0.00695432274379643
|
|
TF
|
TF:M12163_1
|
Factor: KLF3; motif: GRCCRCGCCCN; match class: 1
|
0.0280686118002343
|
89
|
66
|
3
|
0.0454545454545455
|
0.0337078651685393
|
|
TF
|
TF:M10849
|
Factor: Cdx-2; motif: NRTCGTAANNNN
|
0.0285239719009816
|
11163
|
10
|
10
|
1
|
0.000895816536773269
|
|
TF
|
TF:M10172
|
Factor: HKR2; motif: NGNCNGAGGGMGGAGG
|
0.0287629016716216
|
1391
|
22
|
6
|
0.272727272727273
|
0.00431344356578001
|
|
TF
|
TF:M06845
|
Factor: ZBTB38; motif: NTYRATGCAMY
|
0.028787434068377
|
153
|
39
|
3
|
0.0769230769230769
|
0.0196078431372549
|
|
TF
|
TF:M02015
|
Factor: HNF-3gamma; motif: TGTTTRYT
|
0.0290448286346597
|
2809
|
32
|
11
|
0.34375
|
0.00391598433606266
|
|
TF
|
TF:M04932
|
Factor: Sp2; motif: AAGGGGCGG
|
0.0294791010614796
|
2593
|
81
|
20
|
0.246913580246914
|
0.00771307365985345
|
|
TF
|
TF:M11918
|
Factor: Brn-4; motif: NTATGCWAATKAG
|
0.0296654204766829
|
77
|
78
|
3
|
0.0384615384615385
|
0.038961038961039
|
|
TF
|
TF:M11490
|
Factor: LBP-1; motif: NRCCGGTNNNNACCGGYN
|
0.0297033706331768
|
6748
|
86
|
42
|
0.488372093023256
|
0.00622406639004149
|
|
TF
|
TF:M09637
|
Factor: Mef-2a; motif: NKCTATTTTTRGM
|
0.0299270958457964
|
1238
|
11
|
4
|
0.363636363636364
|
0.00323101777059774
|
|
TF
|
TF:M11296
|
Factor: jdp2; motif: NRTGAYGTCAYN
|
0.0303817357600057
|
2950
|
72
|
20
|
0.277777777777778
|
0.00677966101694915
|
|
TF
|
TF:M00040_1
|
Factor: ATF-2; motif: TTACGTAA; match class: 1
|
0.0305863493945109
|
6460
|
29
|
17
|
0.586206896551724
|
0.00263157894736842
|
|
TF
|
TF:M00378
|
Factor: Pax-4; motif: NNNNNYCACCCB
|
0.030729990781867
|
17858
|
98
|
96
|
0.979591836734694
|
0.00537574196438571
|
|
TF
|
TF:M11280
|
Factor: FOSB; motif: NATGAYGTCATN
|
0.030729990781867
|
436
|
47
|
5
|
0.106382978723404
|
0.0114678899082569
|
|
TF
|
TF:M06156
|
Factor: ZNF75; motif: NCATSCGGCTRC
|
0.0307899384043551
|
313
|
65
|
5
|
0.0769230769230769
|
0.0159744408945687
|
|
TF
|
TF:M01822_1
|
Factor: CPBP; motif: SNCCCNN; match class: 1
|
0.031017677766308
|
18663
|
101
|
101
|
1
|
0.00541177731340085
|
|
TF
|
TF:M01177
|
Factor: SREBP-2; motif: NNGYCACNNSMN
|
0.031017677766308
|
8829
|
95
|
56
|
0.589473684210526
|
0.00634273417148035
|
|
TF
|
TF:M11527
|
Factor: E2F-3; motif: NTTTTGGCGCCAAAAN
|
0.0313211302019605
|
6929
|
101
|
49
|
0.485148514851485
|
0.00707172752200895
|
|
TF
|
TF:M07249_1
|
Factor: ctcf; motif: CCNCNAGRKGGCRSTN; match class: 1
|
0.0316067987613593
|
2074
|
74
|
16
|
0.216216216216216
|
0.0077145612343298
|
|
TF
|
TF:M03966
|
Factor: ELF1; motif: AACCCGGAAGTR
|
0.0317182062646773
|
6610
|
33
|
19
|
0.575757575757576
|
0.00287443267776097
|
|
TF
|
TF:M09877_1
|
Factor: BMAL1; motif: NSCACGTGNSN; match class: 1
|
0.0317182062646773
|
222
|
92
|
5
|
0.0543478260869565
|
0.0225225225225225
|
|
TF
|
TF:M11304_1
|
Factor: batf3; motif: NRTGAYGTCAYN; match class: 1
|
0.0320464994254033
|
730
|
29
|
5
|
0.172413793103448
|
0.00684931506849315
|
|
TF
|
TF:M11225
|
Factor: CREM; motif: NNTGACGTCANN
|
0.0321546589941267
|
2710
|
29
|
10
|
0.344827586206897
|
0.003690036900369
|
|
TF
|
TF:M03967
|
Factor: ELF3; motif: WACCCGGAAGTAN
|
0.032288881607416
|
7610
|
69
|
38
|
0.550724637681159
|
0.0049934296977661
|
|
TF
|
TF:M11224_1
|
Factor: CREM; motif: NRTGATGTCAYN; match class: 1
|
0.032288881607416
|
1025
|
72
|
10
|
0.138888888888889
|
0.00975609756097561
|
|
TF
|
TF:M11306_1
|
Factor: ATF-2; motif: NRTGAYGTMAYN; match class: 1
|
0.032288881607416
|
1025
|
72
|
10
|
0.138888888888889
|
0.00975609756097561
|
|
TF
|
TF:M12170
|
Factor: AP-2rep; motif: NRCCACGCCCN
|
0.0323768553439566
|
212
|
60
|
4
|
0.0666666666666667
|
0.0188679245283019
|
|
TF
|
TF:M10550_1
|
Factor: ZNF684; motif: MAAGGGGTGGACTGT; match class: 1
|
0.0324909702763113
|
9336
|
87
|
54
|
0.620689655172414
|
0.0057840616966581
|
|
TF
|
TF:M11323
|
Factor: C/EBPgamma; motif: NRTTGCGYAANN
|
0.0328569452603668
|
1237
|
70
|
11
|
0.157142857142857
|
0.00889248181083266
|
|
TF
|
TF:M03572
|
Factor: DEC1; motif: CNCACRTGASC
|
0.0329501883372214
|
6713
|
24
|
15
|
0.625
|
0.00223447043050797
|
|
TF
|
TF:M01241
|
Factor: BEN; motif: CWGCGAYA
|
0.032969496446588
|
7479
|
24
|
16
|
0.666666666666667
|
0.00213932343896243
|
|
TF
|
TF:M00007
|
Factor: Elk-1; motif: NAAACMGGAAGTNCVH
|
0.033030713485156
|
5873
|
11
|
8
|
0.727272727272727
|
0.00136216584369147
|
|
TF
|
TF:M12166_1
|
Factor: CPBP; motif: NGCCACGCCCN; match class: 1
|
0.0331483231593914
|
162
|
79
|
4
|
0.0506329113924051
|
0.0246913580246914
|
|
TF
|
TF:M00737_1
|
Factor: E2F-1:DP-2; motif: TTTSSCGC; match class: 1
|
0.0332208475030233
|
2933
|
88
|
23
|
0.261363636363636
|
0.0078418002045687
|
|
TF
|
TF:M09603_1
|
Factor: Egr-1; motif: NNNNNGYGKGGGNGGGNN; match class: 1
|
0.0334050971568037
|
1317
|
95
|
14
|
0.147368421052632
|
0.0106302201974184
|
|
TF
|
TF:M11222
|
Factor: CREB1; motif: NRTGAYGTCAYN
|
0.0335737893931078
|
3688
|
19
|
9
|
0.473684210526316
|
0.00244034707158351
|
|
TF
|
TF:M11254_1
|
Factor: AIBZIP; motif: GRTGAYGTCAYC; match class: 1
|
0.0336232054920418
|
742
|
29
|
5
|
0.172413793103448
|
0.00673854447439353
|
|
TF
|
TF:M07355_1
|
Factor: GR; motif: AGAACAN; match class: 1
|
0.0336232054920418
|
1024
|
7
|
3
|
0.428571428571429
|
0.0029296875
|
|
TF
|
TF:M04519_1
|
Factor: E2F-4; motif: TTTGGCGCCAAA; match class: 1
|
0.0338651260188563
|
2547
|
95
|
22
|
0.231578947368421
|
0.00863761287789556
|
|
TF
|
TF:M10460_1
|
Factor: ZNF273; motif: GAGAGGAGCTAC; match class: 1
|
0.0338651260188563
|
5829
|
27
|
15
|
0.555555555555556
|
0.00257334019557385
|
|
TF
|
TF:M07297_1
|
Factor: MAZ; motif: CCCTCCCYCYN; match class: 1
|
0.0338651260188563
|
685
|
59
|
7
|
0.11864406779661
|
0.0102189781021898
|
|
TF
|
TF:M11305_1
|
Factor: batf3; motif: NRTGACGTCAYN; match class: 1
|
0.0339532123800909
|
745
|
29
|
5
|
0.172413793103448
|
0.00671140939597315
|
|
TF
|
TF:M11305
|
Factor: batf3; motif: NRTGACGTCAYN
|
0.0339532123800909
|
745
|
29
|
5
|
0.172413793103448
|
0.00671140939597315
|
|
TF
|
TF:M11435
|
Factor: Elk-1; motif: NNCCGGAAGTN
|
0.0340515039086337
|
11728
|
75
|
56
|
0.746666666666667
|
0.00477489768076398
|
|
TF
|
TF:M04015
|
Factor: IRF-4; motif: NCGAAACCGAAACYA
|
0.0341485312963011
|
2038
|
44
|
11
|
0.25
|
0.00539744847890088
|
|
TF
|
TF:M07353
|
Factor: E2A; motif: CAGNTGNN
|
0.0344162374988703
|
2645
|
30
|
10
|
0.333333333333333
|
0.00378071833648393
|
|
TF
|
TF:M00327
|
Factor: Pax-3; motif: NNNNNNCGTCACGSTYNNNNN
|
0.0346492851607936
|
13318
|
88
|
71
|
0.806818181818182
|
0.00533113080042048
|
|
TF
|
TF:M05361
|
Factor: Sp6; motif: WGGGCGG
|
0.0346492851607936
|
8578
|
88
|
51
|
0.579545454545455
|
0.00594544182793192
|
|
TF
|
TF:M05444
|
Factor: CPBP; motif: NGGGCGG
|
0.0346492851607936
|
8578
|
88
|
51
|
0.579545454545455
|
0.00594544182793192
|
|
TF
|
TF:M05332
|
Factor: Sp2; motif: WGGGCGG
|
0.0346492851607936
|
8578
|
88
|
51
|
0.579545454545455
|
0.00594544182793192
|
|
TF
|
TF:M00425_1
|
Factor: E2F; motif: TTTCGCGC; match class: 1
|
0.0348229596804004
|
5355
|
89
|
36
|
0.404494382022472
|
0.00672268907563025
|
|
TF
|
TF:M11659
|
Factor: HSF4; motif: NGAANRTTCTAGAAN
|
0.0348229596804004
|
329
|
92
|
6
|
0.0652173913043478
|
0.0182370820668693
|
|
TF
|
TF:M11656
|
Factor: HSF1; motif: RGAANRTTCYRGAAN
|
0.0348229596804004
|
2033
|
97
|
19
|
0.195876288659794
|
0.00934579439252336
|
|
TF
|
TF:M00187_1
|
Factor: USF; motif: GYCACGTGNC; match class: 1
|
0.0348499575188619
|
1528
|
83
|
14
|
0.168674698795181
|
0.00916230366492147
|
|
TF
|
TF:M11270
|
Factor: JunD; motif: NRTGACGTCATS
|
0.0348499575188619
|
9380
|
22
|
17
|
0.772727272727273
|
0.00181236673773987
|
|
TF
|
TF:M11379_1
|
Factor: ESE-1; motif: ACCCGGAAGTRNNNNNNNNWWWW; match class: 1
|
0.0348499575188619
|
396
|
17
|
3
|
0.176470588235294
|
0.00757575757575758
|
|
TF
|
TF:M11417
|
Factor: c-Ets-2; motif: NACCGGAAGYRCTTCCGGTN
|
0.0349027606288739
|
14334
|
92
|
78
|
0.847826086956522
|
0.0054416073670992
|
|
TF
|
TF:M12139
|
Factor: Egr-1; motif: NMCRCCCMCNCNN
|
0.0349027606288739
|
3773
|
82
|
26
|
0.317073170731707
|
0.00689106811555791
|
|
TF
|
TF:M00774
|
Factor: NF-kappaB; motif: NGGGANTTYCCMNNNN
|
0.0356189924856515
|
1818
|
124
|
21
|
0.169354838709677
|
0.0115511551155116
|
|
TF
|
TF:M03907
|
Factor: HINFP; motif: CNRCGTCCGCNN
|
0.0357960279950277
|
1052
|
95
|
12
|
0.126315789473684
|
0.0114068441064639
|
|
TF
|
TF:M12185
|
Factor: BTEB4; motif: RCCACGCCCC
|
0.035963372017396
|
2283
|
81
|
18
|
0.222222222222222
|
0.00788436268068331
|
|
TF
|
TF:M00360
|
Factor: Pax-3; motif: TCGTCACRCTTHM
|
0.0360732801048856
|
12238
|
90
|
68
|
0.755555555555556
|
0.00555646347442393
|
|
TF
|
TF:M05775
|
Factor: ZXDL; motif: NGGGGWS
|
0.0364472798647328
|
6964
|
99
|
48
|
0.484848484848485
|
0.00689259046524986
|
|
TF
|
TF:M03959_1
|
Factor: E2F2; motif: NNTTTTGGCGCCAAAAWN; match class: 1
|
0.0365436861449198
|
2951
|
59
|
17
|
0.288135593220339
|
0.00576075906472382
|
|
TF
|
TF:M03811
|
Factor: AP-2gamma; motif: GCCYNCRGSN
|
0.0365786769563848
|
8852
|
82
|
49
|
0.597560975609756
|
0.00553547220967013
|
|
TF
|
TF:M11270_1
|
Factor: JunD; motif: NRTGACGTCATS; match class: 1
|
0.0366617070638742
|
4298
|
29
|
13
|
0.448275862068966
|
0.00302466263378315
|
|
TF
|
TF:M00114_1
|
Factor: Tax/CREB; motif: GGGGGTTGACGYANA; match class: 1
|
0.0368451261710371
|
1564
|
99
|
16
|
0.161616161616162
|
0.010230179028133
|
|
TF
|
TF:M07349
|
Factor: AP-2gamma; motif: NNNNWGCCYNCRGSCN
|
0.0371893078240146
|
3333
|
3
|
3
|
1
|
0.0009000900090009
|
|
TF
|
TF:M08226
|
Factor: Elk-1:Pax-9; motif: ACCGGAACYACGCWYSANTG
|
0.0371893078240146
|
12039
|
81
|
61
|
0.753086419753086
|
0.00506686601877232
|
|
TF
|
TF:M03946
|
Factor: GRHL1; motif: AACCGGTNNAACCGGTT
|
0.0373799079564268
|
68
|
99
|
3
|
0.0303030303030303
|
0.0441176470588235
|
|
TF
|
TF:M01587
|
Factor: FPM315; motif: SRGGGAGGAGGN
|
0.037761753777056
|
3246
|
18
|
8
|
0.444444444444444
|
0.00246457178065311
|
|
TF
|
TF:M12173_1
|
Factor: GKLF; motif: NNCCMCRCCCN; match class: 1
|
0.0381320162165388
|
4965
|
96
|
36
|
0.375
|
0.00725075528700906
|
|
TF
|
TF:M00724
|
Factor: HNF3alpha; motif: TRTTTGYTYWN
|
0.0381320162165388
|
2368
|
39
|
11
|
0.282051282051282
|
0.00464527027027027
|
|
TF
|
TF:M08457_1
|
Factor: ER71:SREBP-2; motif: NTSACGTGACGGAARY; match class: 1
|
0.0381671086354819
|
5591
|
89
|
37
|
0.415730337078652
|
0.00661777857270613
|
|
TF
|
TF:M03824
|
Factor: GCMb; motif: RCCCKCAT
|
0.0385075685043022
|
1550
|
83
|
14
|
0.168674698795181
|
0.00903225806451613
|
|
TF
|
TF:M08930_1
|
Factor: JUND:FRA-1; motif: RTGACGTMAY; match class: 1
|
0.0385109780408059
|
1607
|
72
|
13
|
0.180555555555556
|
0.00808960796515246
|
|
TF
|
TF:M09862_1
|
Factor: ZNF614; motif: NCYCWGCCYYNNN; match class: 1
|
0.0388235202722389
|
4193
|
124
|
39
|
0.314516129032258
|
0.00930121631290246
|
|
TF
|
TF:M11830_1
|
Factor: LRH-1; motif: TGACCTTGRNYCAAGGTCA; match class: 1
|
0.0388235202722389
|
6100
|
77
|
35
|
0.454545454545455
|
0.00573770491803279
|
|
TF
|
TF:M00056_1
|
Factor: myogenin; motif: CRSCTGTTBNNTTTGGCACGSNGCCARCH; match class: 1
|
0.0388235202722389
|
5754
|
101
|
42
|
0.415841584158416
|
0.0072992700729927
|
|
TF
|
TF:M01227
|
Factor: MafB; motif: GNTGAC
|
0.0390528791454468
|
11727
|
95
|
69
|
0.726315789473684
|
0.00588385776413405
|
|
TF
|
TF:M01253_1
|
Factor: CNOT3; motif: GGCCGCGSSS; match class: 1
|
0.0390528791454468
|
1026
|
124
|
14
|
0.112903225806452
|
0.01364522417154
|
|
TF
|
TF:M03878
|
Factor: MIBP1; motif: WNWCCCCAGCTR
|
0.0392478838908522
|
2793
|
78
|
20
|
0.256410256410256
|
0.00716075904045829
|
|
TF
|
TF:M12005
|
Factor: GMEB-1; motif: KNACGTNRNNACGTAN
|
0.0397468202589004
|
1166
|
56
|
9
|
0.160714285714286
|
0.00771869639794168
|
|
TF
|
TF:M11247
|
Factor: Luman; motif: YGCCACGTCAYC
|
0.0397525439139942
|
174
|
79
|
4
|
0.0506329113924051
|
0.0229885057471264
|
|
TF
|
TF:M00122
|
Factor: USF; motif: NNRNCACGTGNYNN
|
0.0400279958593133
|
2413
|
72
|
17
|
0.236111111111111
|
0.00704517198508081
|
|
TF
|
TF:M00122_1
|
Factor: USF; motif: NNRNCACGTGNYNN; match class: 1
|
0.0400279958593133
|
2413
|
72
|
17
|
0.236111111111111
|
0.00704517198508081
|
|
TF
|
TF:M10024_1
|
Factor: p63; motif: NRCAWGYCCAGRCWTGYNN; match class: 1
|
0.0401434198745545
|
326
|
68
|
5
|
0.0735294117647059
|
0.0153374233128834
|
|
TF
|
TF:M10116
|
Factor: ZID; motif: GGNGMTGGAGCCN
|
0.0402096528553071
|
1601
|
81
|
14
|
0.172839506172839
|
0.00874453466583385
|
|
TF
|
TF:M11484_1
|
Factor: LBP-32; motif: NAACCGGTTN; match class: 1
|
0.0402096528553071
|
129
|
107
|
4
|
0.0373831775700935
|
0.0310077519379845
|
|
TF
|
TF:M12187
|
Factor: KLF17; motif: NMCCACNCWCCCNTN
|
0.0402096528553071
|
2886
|
6
|
4
|
0.666666666666667
|
0.00138600138600139
|
|
TF
|
TF:M11484
|
Factor: LBP-32; motif: NAACCGGTTN
|
0.0402096528553071
|
129
|
107
|
4
|
0.0373831775700935
|
0.0310077519379845
|
|
TF
|
TF:M09972_1
|
Factor: BTEB2; motif: WGGGTGKGGCNGGN; match class: 1
|
0.0403238501594117
|
3919
|
100
|
31
|
0.31
|
0.00791018116866548
|
|
TF
|
TF:M00431
|
Factor: E2F-1; motif: TTTSGCGS
|
0.0403238501594117
|
7257
|
89
|
45
|
0.50561797752809
|
0.00620090946672179
|
|
TF
|
TF:M08220_1
|
Factor: Elk-1:HOXA3; motif: ACCGGWAATKRNNTNWCNNATTAN; match class: 1
|
0.0403979829182593
|
5350
|
4
|
4
|
1
|
0.000747663551401869
|
|
TF
|
TF:M08261
|
Factor: ER71:HOXB13; motif: NNCGGAWGTNGTWAAN
|
0.0405833293168834
|
319
|
99
|
6
|
0.0606060606060606
|
0.0188087774294671
|
|
TF
|
TF:M03866_1
|
Factor: c-Fos:c-Jun; motif: NTGACTCAN; match class: 1
|
0.0408629089617929
|
89
|
79
|
3
|
0.0379746835443038
|
0.0337078651685393
|
|
TF
|
TF:M11018_1
|
Factor: IRX-1; motif: NACRYNNNNNNNNRYGNN; match class: 1
|
0.0409289762463059
|
15920
|
90
|
82
|
0.911111111111111
|
0.00515075376884422
|
|
TF
|
TF:M08947
|
Factor: C-FOS; motif: NNNKATGACGTCATNNN
|
0.0411983297502623
|
7042
|
10
|
8
|
0.8
|
0.00113604089747231
|
|
TF
|
TF:M05617
|
Factor: ZNF500; motif: NGTTCCGGGGRW
|
0.0412670659158066
|
59
|
120
|
3
|
0.025
|
0.0508474576271186
|
|
TF
|
TF:M08213
|
Factor: Elk-1:TBR2; motif: TNRCACCGGAAGN
|
0.0417098064865694
|
6273
|
26
|
15
|
0.576923076923077
|
0.00239120038259206
|
|
TF
|
TF:M04617
|
Factor: LRF; motif: NGNAGNGGGTYN
|
0.0419881138044728
|
6164
|
82
|
37
|
0.451219512195122
|
0.00600259571706684
|
|
TF
|
TF:M04012
|
Factor: HSFY1; motif: TTTCGAACG
|
0.0424681470134645
|
1013
|
65
|
9
|
0.138461538461538
|
0.00888450148075025
|
|
TF
|
TF:M08891
|
Factor: NFKAPPAB; motif: NNGGGANTTCCCN
|
0.0425252177394613
|
2110
|
12
|
5
|
0.416666666666667
|
0.0023696682464455
|
|
TF
|
TF:M12187_1
|
Factor: KLF17; motif: NMCCACNCWCCCNTN; match class: 1
|
0.0425982403010734
|
377
|
38
|
4
|
0.105263157894737
|
0.0106100795755968
|
|
TF
|
TF:M08885_1
|
Factor: HAIRYLIKE; motif: NNNNCANGTG; match class: 1
|
0.0428722508717282
|
2110
|
96
|
19
|
0.197916666666667
|
0.00900473933649289
|
|
TF
|
TF:M01138
|
Factor: RORalpha; motif: WAANTRGGTCA
|
0.0430135118669243
|
407
|
56
|
5
|
0.0892857142857143
|
0.0122850122850123
|
|
TF
|
TF:M11987
|
Factor: NFATc3; motif: NAYGGAAANW
|
0.0431056448551638
|
2409
|
24
|
8
|
0.333333333333333
|
0.0033208800332088
|
|
TF
|
TF:M11222_1
|
Factor: CREB1; motif: NRTGAYGTCAYN; match class: 1
|
0.0434691332926361
|
1088
|
72
|
10
|
0.138888888888889
|
0.00919117647058824
|
|
TF
|
TF:M10550
|
Factor: ZNF684; motif: MAAGGGGTGGACTGT
|
0.0434691332926361
|
15246
|
69
|
62
|
0.898550724637681
|
0.00406664043027679
|
|
TF
|
TF:M08870_1
|
Factor: DEC1; motif: NCNCACRTGNSC; match class: 1
|
0.0436110381971473
|
2440
|
7
|
4
|
0.571428571428571
|
0.00163934426229508
|
|
TF
|
TF:M06537
|
Factor: CTCFL; motif: NSGTGGGCGG
|
0.0436474767410619
|
746
|
88
|
9
|
0.102272727272727
|
0.0120643431635389
|
|
TF
|
TF:M01023_1
|
Factor: HSF1; motif: NTTCTRGAAVNTTCTYM; match class: 1
|
0.0436497139779916
|
254
|
29
|
3
|
0.103448275862069
|
0.0118110236220472
|
|
TF
|
TF:M07424
|
Factor: MEF-2; motif: NNYTATTTTTAGN
|
0.0436497139779916
|
704
|
11
|
3
|
0.272727272727273
|
0.00426136363636364
|
|
TF
|
TF:M04571
|
Factor: TFEC; motif: RNCACRTGAY
|
0.044270102678108
|
1645
|
72
|
13
|
0.180555555555556
|
0.00790273556231003
|
|
TF
|
TF:M03948_1
|
Factor: TFCP2; motif: AAACCGGTTY; match class: 1
|
0.0442843962974642
|
134
|
107
|
4
|
0.0373831775700935
|
0.0298507462686567
|
|
TF
|
TF:M02055
|
Factor: ESE-1; motif: NSMGGAARTN
|
0.0442962094053087
|
7559
|
93
|
48
|
0.516129032258065
|
0.00635004630242096
|
|
TF
|
TF:M01214
|
Factor: ESE-1; motif: NSMGGAARTN
|
0.0442962094053087
|
7559
|
93
|
48
|
0.516129032258065
|
0.00635004630242096
|
|
TF
|
TF:M01241_1
|
Factor: BEN; motif: CWGCGAYA; match class: 1
|
0.0442962094053087
|
2097
|
22
|
7
|
0.318181818181818
|
0.0033381020505484
|
|
TF
|
TF:M11289_1
|
Factor: ATF-3; motif: NRTGACGTCAYN; match class: 1
|
0.0446089249757998
|
1661
|
34
|
8
|
0.235294117647059
|
0.00481637567730283
|
|
TF
|
TF:M04691
|
Factor: Kaiso; motif: TCTCGCGAG
|
0.0446584661604958
|
15113
|
84
|
74
|
0.880952380952381
|
0.00489644676768345
|
|
TF
|
TF:M12186_1
|
Factor: BTEB4; motif: NCCACGCCCM; match class: 1
|
0.0446584661604958
|
3958
|
96
|
30
|
0.3125
|
0.00757958564931784
|
|
TF
|
TF:M01002
|
Factor: DEAF1; motif: RNNNRTTCGGGNRTTTCCGGRRNKN
|
0.0446584661604958
|
1086
|
84
|
11
|
0.130952380952381
|
0.0101289134438306
|
|
TF
|
TF:M08225
|
Factor: Elk-1:Pax-5; motif: ACCGGAACYACGCWTSANYG
|
0.0446584661604958
|
14039
|
98
|
81
|
0.826530612244898
|
0.00576964171237268
|
|
TF
|
TF:M10100
|
Factor: TFEA; motif: RGTCACGTGA
|
0.0449411588746926
|
6114
|
34
|
18
|
0.529411764705882
|
0.00294406280667321
|
|
TF
|
TF:M10058_1
|
Factor: Six-2; motif: KGAAAYNTGAKMC; match class: 1
|
0.0454405103321711
|
75
|
99
|
3
|
0.0303030303030303
|
0.04
|
|
TF
|
TF:M11880
|
Factor: pax-6; motif: NYACGCWTSANYGMNCN
|
0.0458367539838368
|
13087
|
90
|
71
|
0.788888888888889
|
0.00542523114541148
|
|
TF
|
TF:M04064_1
|
Factor: Pax-5; motif: NGTCACGCWTSANTGMNY; match class: 1
|
0.0462136460208557
|
3538
|
28
|
11
|
0.392857142857143
|
0.00310910118711136
|
|
TF
|
TF:M08957
|
Factor: NURR1:RXR-ALPHA; motif: NRGGTCRTTGACCYN
|
0.046375219674974
|
8851
|
83
|
49
|
0.590361445783133
|
0.00553609761608858
|
|
TF
|
TF:M09814
|
Factor: MYCN; motif: NNCCACGTGGNN
|
0.046375219674974
|
2066
|
92
|
18
|
0.195652173913043
|
0.00871248789932236
|
|
TF
|
TF:M09814_1
|
Factor: MYCN; motif: NNCCACGTGGNN; match class: 1
|
0.046375219674974
|
2066
|
92
|
18
|
0.195652173913043
|
0.00871248789932236
|
|
TF
|
TF:M04595
|
Factor: SALL2; motif: GGGTGGG
|
0.046375219674974
|
13013
|
96
|
75
|
0.78125
|
0.00576346730192884
|
|
TF
|
TF:M09595
|
Factor: BCL-11A; motif: NAAAGAGGAAGTGARAN
|
0.0464240984405675
|
3696
|
3
|
3
|
1
|
0.000811688311688312
|
|
TF
|
TF:M08295
|
Factor: FOXO1A:Elf-1; motif: NAGAAAACCGAANM
|
0.0464240984405675
|
11711
|
64
|
48
|
0.75
|
0.00409871061395269
|
|
TF
|
TF:M04008
|
Factor: HSF1; motif: TTCTAGAAYNTTC
|
0.0466240154719352
|
358
|
92
|
6
|
0.0652173913043478
|
0.0167597765363128
|
|
TF
|
TF:M00428_1
|
Factor: E2F-1; motif: NKTSSCGC; match class: 1
|
0.0468435582980963
|
3615
|
92
|
27
|
0.293478260869565
|
0.00746887966804979
|
|
TF
|
TF:M00920
|
Factor: E2F; motif: NKCGCGCSAAAN
|
0.0472447272035209
|
5663
|
75
|
32
|
0.426666666666667
|
0.00565071516863853
|
|
TF
|
TF:M01660
|
Factor: GABP-alpha; motif: CTTCCK
|
0.0478402411867286
|
11116
|
70
|
50
|
0.714285714285714
|
0.00449802087081684
|
|
TF
|
TF:M01863
|
Factor: ATF-3; motif: RTKRCGTCANN
|
0.048095298621889
|
1885
|
19
|
6
|
0.315789473684211
|
0.00318302387267905
|
|
TF
|
TF:M00260_1
|
Factor: HLF; motif: GTTACRYAAT; match class: 1
|
0.0483694475863588
|
465
|
17
|
3
|
0.176470588235294
|
0.00645161290322581
|
|
TF
|
TF:M08825_1
|
Factor: NF-1B; motif: KCCAGANWN; match class: 1
|
0.0486436093761575
|
1081
|
52
|
8
|
0.153846153846154
|
0.00740055504162812
|
|
TF
|
TF:M12285
|
Factor: ZNF647; motif: NTAGGCCTAN
|
0.0490762285803769
|
1290
|
44
|
8
|
0.181818181818182
|
0.0062015503875969
|
|
TF
|
TF:M08739
|
Factor: rfx3:FIGLA; motif: TRGCAACNNNMCASSTGKN
|
0.0495548796036356
|
112
|
69
|
3
|
0.0434782608695652
|
0.0267857142857143
|
|
TF
|
TF:M01770_1
|
Factor: XBP-1; motif: WNNGMCACGTC; match class: 1
|
0.0499653539522949
|
4554
|
92
|
32
|
0.347826086956522
|
0.00702678963548529
|
|
WP
|
WP:WP366
|
TGF-beta Signaling Pathway
|
1.79327749913997e-07
|
133
|
8
|
6
|
0.75
|
0.0451127819548872
|
|
WP
|
WP:WP4754
|
IL-18 signaling pathway
|
1.91672354513517e-07
|
279
|
58
|
15
|
0.258620689655172
|
0.0537634408602151
|
|
WP
|
WP:WP2431
|
Spinal Cord Injury
|
1.65755639512521e-06
|
118
|
13
|
6
|
0.461538461538462
|
0.0508474576271186
|
|
WP
|
WP:WP2374
|
Oncostatin M Signaling Pathway
|
2.73816426047193e-05
|
66
|
35
|
6
|
0.171428571428571
|
0.0909090909090909
|
|
WP
|
WP:WP3611
|
Photodynamic therapy-induced AP-1 survival signaling.
|
2.73816426047193e-05
|
51
|
25
|
5
|
0.2
|
0.0980392156862745
|
|
WP
|
WP:WP2355
|
Corticotropin-releasing hormone signaling pathway
|
2.85770729752029e-05
|
91
|
44
|
7
|
0.159090909090909
|
0.0769230769230769
|
|
WP
|
WP:WP3527
|
Preimplantation Embryo
|
2.85770729752029e-05
|
60
|
23
|
5
|
0.217391304347826
|
0.0833333333333333
|
|
WP
|
WP:WP4877
|
Host-pathogen interaction of human corona viruses - MAPK signaling
|
6.16085007946574e-05
|
36
|
6
|
3
|
0.5
|
0.0833333333333333
|
|
WP
|
WP:WP4659
|
Gastrin Signaling Pathway
|
0.000324001400376791
|
114
|
35
|
6
|
0.171428571428571
|
0.0526315789473684
|
|
WP
|
WP:WP2032
|
Human Thyroid Stimulating Hormone (TSH) signaling pathway
|
0.000342660988500174
|
68
|
6
|
3
|
0.5
|
0.0441176470588235
|
|
WP
|
WP:WP4919
|
Neuroinflammation
|
0.000574615846643826
|
13
|
35
|
3
|
0.0857142857142857
|
0.230769230769231
|
|
WP
|
WP:WP2882
|
Nuclear Receptors Meta-Pathway
|
0.00061879772518157
|
321
|
31
|
8
|
0.258064516129032
|
0.0249221183800623
|
|
WP
|
WP:WP2840
|
Hair Follicle Development: Cytodifferentiation - Part 3 of 3
|
0.00061879772518157
|
89
|
6
|
3
|
0.5
|
0.0337078651685393
|
|
WP
|
WP:WP516
|
Hypertrophy Model
|
0.000629044094861073
|
20
|
25
|
3
|
0.12
|
0.15
|
|
WP
|
WP:WP2873
|
Aryl Hydrocarbon Receptor Pathway
|
0.000662315074295228
|
48
|
31
|
4
|
0.129032258064516
|
0.0833333333333333
|
|
WP
|
WP:WP382
|
MAPK Signaling Pathway
|
0.00143883200405895
|
249
|
24
|
6
|
0.25
|
0.0240963855421687
|
|
WP
|
WP:WP2380
|
Brain-Derived Neurotrophic Factor (BDNF) signaling pathway
|
0.00165853766839251
|
144
|
6
|
3
|
0.5
|
0.0208333333333333
|
|
WP
|
WP:WP437
|
EGF/EGFR Signaling Pathway
|
0.0019673376388079
|
163
|
6
|
3
|
0.5
|
0.0184049079754601
|
|
WP
|
WP:WP481
|
Insulin Signaling
|
0.0019673376388079
|
161
|
6
|
3
|
0.5
|
0.0186335403726708
|
|
WP
|
WP:WP289
|
Myometrial Relaxation and Contraction Pathways
|
0.00241007953272208
|
155
|
7
|
3
|
0.428571428571429
|
0.0193548387096774
|
|
WP
|
WP:WP2037
|
Prolactin Signaling Pathway
|
0.00297170759064122
|
76
|
96
|
6
|
0.0625
|
0.0789473684210526
|
|
WP
|
WP:WP1772
|
Apoptosis Modulation and Signaling
|
0.00297170759064122
|
93
|
52
|
5
|
0.0961538461538462
|
0.0537634408602151
|
|
WP
|
WP:WP254
|
Apoptosis
|
0.00303059308147176
|
86
|
57
|
5
|
0.087719298245614
|
0.0581395348837209
|
|
WP
|
WP:WP395
|
IL-4 Signaling Pathway
|
0.00439278576156728
|
55
|
99
|
5
|
0.0505050505050505
|
0.0909090909090909
|
|
WP
|
WP:WP3594
|
Circadian rhythm related genes
|
0.00509461680543102
|
206
|
113
|
10
|
0.0884955752212389
|
0.0485436893203883
|
|
WP
|
WP:WP4216
|
Chromosomal and microsatellite instability in colorectal cancer
|
0.00509461680543102
|
74
|
78
|
5
|
0.0641025641025641
|
0.0675675675675676
|
|
WP
|
WP:WP3888
|
VEGFA-VEGFR2 Signaling Pathway
|
0.0054926161013371
|
437
|
85
|
13
|
0.152941176470588
|
0.0297482837528604
|
|
WP
|
WP:WP3614
|
Photodynamic therapy-induced HIF-1 survival signaling
|
0.00976243423892745
|
37
|
52
|
3
|
0.0576923076923077
|
0.0810810810810811
|
|
WP
|
WP:WP4149
|
White fat cell differentiation
|
0.0137899326845454
|
33
|
68
|
3
|
0.0441176470588235
|
0.0909090909090909
|
|
WP
|
WP:WP4262
|
Breast cancer pathway
|
0.0150996149611211
|
156
|
31
|
4
|
0.129032258064516
|
0.0256410256410256
|
|
WP
|
WP:WP4286
|
Genotoxicity pathway
|
0.0229898155412205
|
63
|
88
|
4
|
0.0454545454545455
|
0.0634920634920635
|
|
WP
|
WP:WP236
|
Adipogenesis
|
0.0233543265408951
|
131
|
96
|
6
|
0.0625
|
0.0458015267175573
|
|
WP
|
WP:WP4963
|
p53 transcriptional gene network
|
0.0233894173222047
|
72
|
78
|
4
|
0.0512820512820513
|
0.0555555555555556
|
|
WP
|
WP:WP2516
|
ATM Signaling Pathway
|
0.0276911218670646
|
40
|
78
|
3
|
0.0384615384615385
|
0.075
|
|
WP
|
WP:WP364
|
IL-6 signaling pathway
|
0.029155678821621
|
43
|
75
|
3
|
0.04
|
0.0697674418604651
|
|
WP
|
WP:WP619
|
Type II interferon signaling (IFNG)
|
0.0341381905395858
|
37
|
96
|
3
|
0.03125
|
0.0810810810810811
|
|
WP
|
WP:WP2884
|
NRF2 pathway
|
0.0348851105003574
|
145
|
25
|
3
|
0.12
|
0.0206896551724138
|
|
WP
|
WP:WP4932
|
7q11.23 copy number variation syndrome
|
0.0442243425940301
|
97
|
42
|
3
|
0.0714285714285714
|
0.0309278350515464
|
|
WP
|
WP:WP410
|
Exercise-induced Circadian Regulation
|
0.0460728701498357
|
49
|
85
|
3
|
0.0352941176470588
|
0.0612244897959184
|
|
WP
|
WP:WP4658
|
Small cell lung cancer
|
0.046662039042667
|
98
|
78
|
4
|
0.0512820512820513
|
0.0408163265306122
|