|
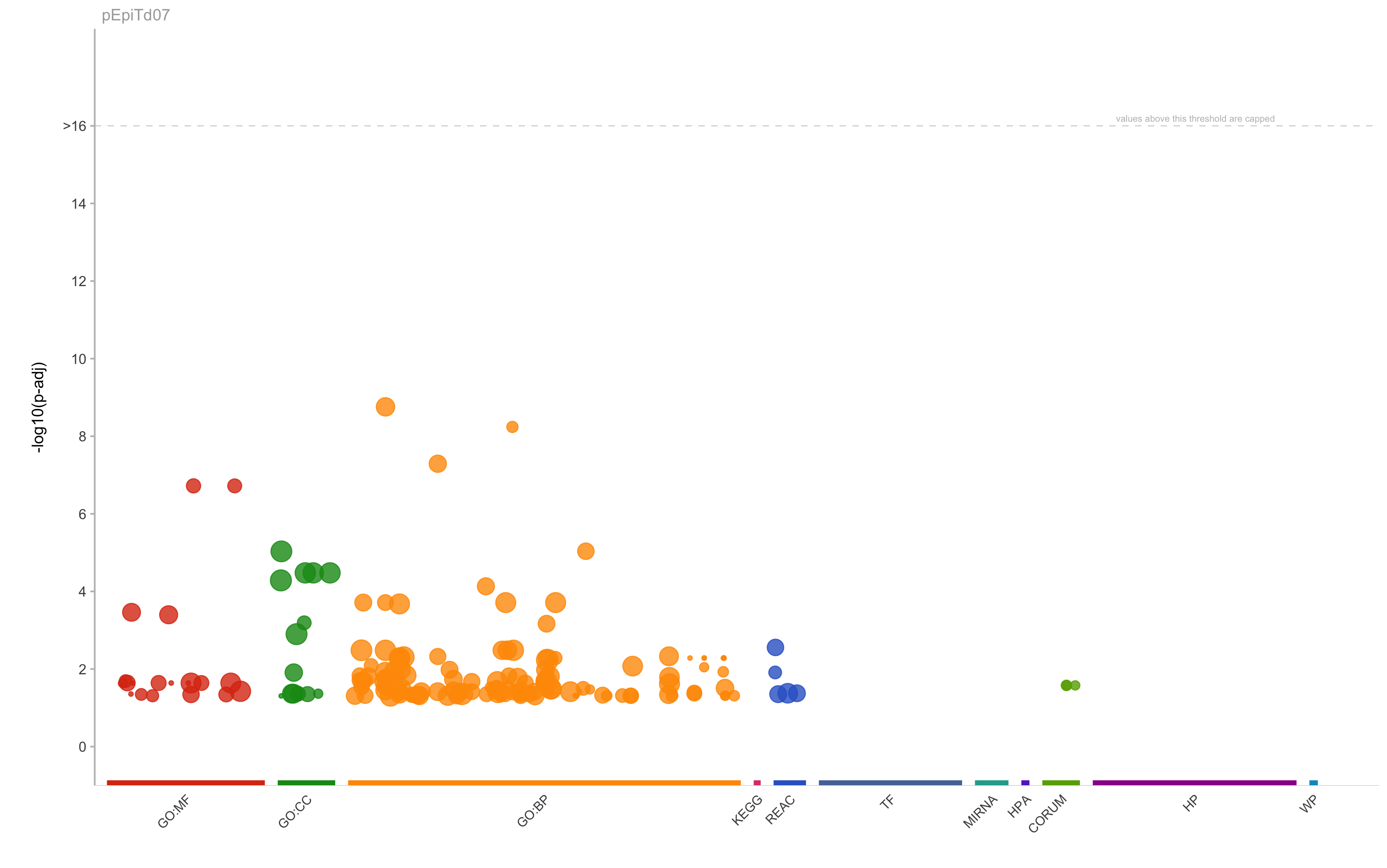
GO:BP
|
GO:0006959
|
humoral immune response
|
1.7469407391308e-09
|
384
|
18
|
10
|
0.555555555555556
|
0.0260416666666667
|
|
GO:BP
|
GO:0044278
|
cell wall disruption in other organism
|
5.75226878917038e-09
|
4
|
3
|
3
|
1
|
0.75
|
|
GO:BP
|
GO:0019730
|
antimicrobial humoral response
|
5.1067504754438e-08
|
145
|
18
|
7
|
0.388888888888889
|
0.0482758620689655
|
|
GO:BP
|
GO:0061844
|
antimicrobial humoral immune response mediated by antimicrobial peptide
|
9.1936865487869e-06
|
81
|
18
|
5
|
0.277777777777778
|
0.0617283950617284
|
|
GO:BP
|
GO:0035821
|
modulation of process of other organism
|
7.38344688717799e-05
|
128
|
18
|
5
|
0.277777777777778
|
0.0390625
|
|
GO:BP
|
GO:0002526
|
acute inflammatory response
|
0.000193768803011602
|
114
|
109
|
8
|
0.073394495412844
|
0.0701754385964912
|
|
GO:BP
|
GO:0051707
|
response to other organism
|
0.000193768803011602
|
1592
|
18
|
10
|
0.555555555555556
|
0.00628140703517588
|
|
GO:BP
|
GO:0043207
|
response to external biotic stimulus
|
0.000193768803011602
|
1594
|
18
|
10
|
0.555555555555556
|
0.00627352572145546
|
|
GO:BP
|
GO:0006953
|
acute-phase response
|
0.000193768803011602
|
50
|
109
|
6
|
0.055045871559633
|
0.12
|
|
GO:BP
|
GO:0009607
|
response to biotic stimulus
|
0.000209920602228183
|
1626
|
18
|
10
|
0.555555555555556
|
0.00615006150061501
|
|
GO:BP
|
GO:0050830
|
defense response to Gram-positive bacterium
|
0.000679318182624963
|
101
|
18
|
4
|
0.222222222222222
|
0.0396039603960396
|
|
GO:BP
|
GO:0002376
|
immune system process
|
0.00328643980275413
|
3278
|
19
|
12
|
0.631578947368421
|
0.0036607687614399
|
|
GO:BP
|
GO:0006955
|
immune response
|
0.00328643980275413
|
2311
|
18
|
10
|
0.555555555555556
|
0.0043271311120727
|
|
GO:BP
|
GO:0042742
|
defense response to bacterium
|
0.00328643980275413
|
349
|
18
|
5
|
0.277777777777778
|
0.0143266475644699
|
|
GO:BP
|
GO:0044419
|
biological process involved in interspecies interaction between organisms
|
0.00328643980275413
|
2255
|
18
|
10
|
0.555555555555556
|
0.00443458980044346
|
|
GO:BP
|
GO:0043434
|
response to peptide hormone
|
0.00328643980275413
|
462
|
3
|
3
|
1
|
0.00649350649350649
|
|
GO:BP
|
GO:1901652
|
response to peptide
|
0.00469132625514894
|
551
|
3
|
3
|
1
|
0.00544464609800363
|
|
GO:BP
|
GO:0019731
|
antibacterial humoral response
|
0.00475366337174542
|
62
|
18
|
3
|
0.166666666666667
|
0.0483870967741935
|
|
GO:BP
|
GO:0010033
|
response to organic substance
|
0.00493829493141793
|
3412
|
22
|
13
|
0.590909090909091
|
0.00381008206330598
|
|
GO:BP
|
GO:0009617
|
response to bacterium
|
0.00521030842241595
|
743
|
18
|
6
|
0.333333333333333
|
0.00807537012113055
|
|
GO:BP
|
GO:0050829
|
defense response to Gram-negative bacterium
|
0.00521030842241595
|
88
|
84
|
5
|
0.0595238095238095
|
0.0568181818181818
|
|
GO:BP
|
GO:0009605
|
response to external stimulus
|
0.00521030842241595
|
2944
|
19
|
11
|
0.578947368421053
|
0.00373641304347826
|
|
GO:BP
|
GO:0050896
|
response to stimulus
|
0.0058937412211612
|
9407
|
82
|
60
|
0.731707317073171
|
0.00637822897842033
|
|
GO:BP
|
GO:0098542
|
defense response to other organism
|
0.00842288124704723
|
1218
|
18
|
7
|
0.388888888888889
|
0.00574712643678161
|
|
GO:BP
|
GO:0009725
|
response to hormone
|
0.0103685587657736
|
921
|
3
|
3
|
1
|
0.00325732899022801
|
|
GO:BP
|
GO:0030449
|
regulation of complement activation
|
0.0104393723083538
|
110
|
17
|
3
|
0.176470588235294
|
0.0272727272727273
|
|
GO:BP
|
GO:0008284
|
positive regulation of cell population proliferation
|
0.0116037907638868
|
982
|
3
|
3
|
1
|
0.00305498981670061
|
|
GO:BP
|
GO:0006952
|
defense response
|
0.0122293662257876
|
1818
|
18
|
8
|
0.444444444444444
|
0.0044004400440044
|
|
GO:BP
|
GO:0010243
|
response to organonitrogen compound
|
0.0145377137044276
|
1085
|
3
|
3
|
1
|
0.00276497695852535
|
|
GO:BP
|
GO:0002251
|
organ or tissue specific immune response
|
0.0147989125289759
|
42
|
51
|
3
|
0.0588235294117647
|
0.0714285714285714
|
|
GO:BP
|
GO:0043616
|
keratinocyte proliferation
|
0.0149444487680458
|
50
|
109
|
4
|
0.036697247706422
|
0.08
|
|
GO:BP
|
GO:0002920
|
regulation of humoral immune response
|
0.0154491374197661
|
133
|
17
|
3
|
0.176470588235294
|
0.0225563909774436
|
|
GO:BP
|
GO:0051094
|
positive regulation of developmental process
|
0.0154707160779863
|
1341
|
89
|
17
|
0.191011235955056
|
0.0126771066368382
|
|
GO:BP
|
GO:1901698
|
response to nitrogen compound
|
0.0163768524375443
|
1172
|
3
|
3
|
1
|
0.00255972696245734
|
|
GO:BP
|
GO:0045087
|
innate immune response
|
0.0171073890154191
|
998
|
18
|
6
|
0.333333333333333
|
0.00601202404809619
|
|
GO:BP
|
GO:0006958
|
complement activation, classical pathway
|
0.0172024960583688
|
142
|
17
|
3
|
0.176470588235294
|
0.0211267605633803
|
|
GO:BP
|
GO:0031099
|
regeneration
|
0.0182959599409618
|
206
|
81
|
6
|
0.0740740740740741
|
0.029126213592233
|
|
GO:BP
|
GO:0002675
|
positive regulation of acute inflammatory response
|
0.0195709997626268
|
29
|
87
|
3
|
0.0344827586206897
|
0.103448275862069
|
|
GO:BP
|
GO:0050793
|
regulation of developmental process
|
0.0204406777197172
|
2605
|
82
|
24
|
0.292682926829268
|
0.00921305182341651
|
|
GO:BP
|
GO:0002455
|
humoral immune response mediated by circulating immunoglobulin
|
0.0205687268933328
|
156
|
17
|
3
|
0.176470588235294
|
0.0192307692307692
|
|
GO:BP
|
GO:0033627
|
cell adhesion mediated by integrin
|
0.0210806185413514
|
73
|
36
|
3
|
0.0833333333333333
|
0.0410958904109589
|
|
GO:BP
|
GO:0006950
|
response to stress
|
0.0210806185413514
|
4170
|
20
|
12
|
0.6
|
0.00287769784172662
|
|
GO:BP
|
GO:0042127
|
regulation of cell population proliferation
|
0.0211218230733968
|
1774
|
5
|
4
|
0.8
|
0.00225479143179256
|
|
GO:BP
|
GO:0050673
|
epithelial cell proliferation
|
0.0224768024282264
|
460
|
109
|
10
|
0.0917431192660551
|
0.0217391304347826
|
|
GO:BP
|
GO:0045995
|
regulation of embryonic development
|
0.0231582984003292
|
63
|
44
|
3
|
0.0681818181818182
|
0.0476190476190476
|
|
GO:BP
|
GO:1901700
|
response to oxygen-containing compound
|
0.0239477986952203
|
1739
|
122
|
24
|
0.19672131147541
|
0.0138010350776308
|
|
GO:BP
|
GO:0006956
|
complement activation
|
0.0272100736183509
|
180
|
17
|
3
|
0.176470588235294
|
0.0166666666666667
|
|
GO:BP
|
GO:0008283
|
cell population proliferation
|
0.0309513212019733
|
2054
|
5
|
4
|
0.8
|
0.00194741966893866
|
|
GO:BP
|
GO:2000027
|
regulation of animal organ morphogenesis
|
0.0309513212019733
|
184
|
106
|
6
|
0.0566037735849057
|
0.0326086956521739
|
|
GO:BP
|
GO:0009719
|
response to endogenous stimulus
|
0.0318855477591047
|
1683
|
3
|
3
|
1
|
0.0017825311942959
|
|
GO:BP
|
GO:0051239
|
regulation of multicellular organismal process
|
0.0322289379645814
|
2861
|
88
|
26
|
0.295454545454545
|
0.00908773156239077
|
|
GO:BP
|
GO:0051241
|
negative regulation of multicellular organismal process
|
0.0329071476500291
|
1160
|
57
|
11
|
0.192982456140351
|
0.00948275862068966
|
|
GO:BP
|
GO:0006954
|
inflammatory response
|
0.0351049960711832
|
794
|
109
|
13
|
0.119266055045872
|
0.0163727959697733
|
|
GO:BP
|
GO:0060429
|
epithelium development
|
0.0383797257555415
|
1325
|
111
|
18
|
0.162162162162162
|
0.0135849056603774
|
|
GO:BP
|
GO:0046849
|
bone remodeling
|
0.0383797257555415
|
93
|
89
|
4
|
0.0449438202247191
|
0.043010752688172
|
|
GO:BP
|
GO:0019724
|
B cell mediated immunity
|
0.0383797257555415
|
226
|
17
|
3
|
0.176470588235294
|
0.0132743362831858
|
|
GO:BP
|
GO:0016064
|
immunoglobulin mediated immune response
|
0.0383797257555415
|
223
|
17
|
3
|
0.176470588235294
|
0.0134529147982063
|
|
GO:BP
|
GO:0045597
|
positive regulation of cell differentiation
|
0.0393445321182609
|
879
|
89
|
12
|
0.134831460674157
|
0.0136518771331058
|
|
GO:BP
|
GO:0042221
|
response to chemical
|
0.0393445321182609
|
4781
|
22
|
13
|
0.590909090909091
|
0.0027190964233424
|
|
GO:BP
|
GO:0032501
|
multicellular organismal process
|
0.043537307263252
|
7946
|
90
|
54
|
0.6
|
0.00679587213692424
|
|
GO:BP
|
GO:0046329
|
negative regulation of JNK cascade
|
0.0444427752751463
|
39
|
106
|
3
|
0.0283018867924528
|
0.0769230769230769
|
|
GO:BP
|
GO:1901617
|
organic hydroxy compound biosynthetic process
|
0.0454660282886512
|
254
|
121
|
7
|
0.0578512396694215
|
0.0275590551181102
|
|
GO:BP
|
GO:0010837
|
regulation of keratinocyte proliferation
|
0.046221982595725
|
39
|
109
|
3
|
0.0275229357798165
|
0.0769230769230769
|
|
GO:BP
|
GO:0015718
|
monocarboxylic acid transport
|
0.046221982595725
|
189
|
82
|
5
|
0.0609756097560976
|
0.0264550264550265
|
|
GO:BP
|
GO:0015849
|
organic acid transport
|
0.0482231101761569
|
331
|
122
|
8
|
0.0655737704918033
|
0.0241691842900302
|
|
GO:BP
|
GO:0002673
|
regulation of acute inflammatory response
|
0.0482231101761569
|
50
|
87
|
3
|
0.0344827586206897
|
0.06
|
|
GO:BP
|
GO:0046942
|
carboxylic acid transport
|
0.0487785466817079
|
294
|
82
|
6
|
0.0731707317073171
|
0.0204081632653061
|
|
GO:BP
|
GO:0045616
|
regulation of keratinocyte differentiation
|
0.0487785466817079
|
43
|
109
|
3
|
0.0275229357798165
|
0.0697674418604651
|
|
GO:BP
|
GO:0030155
|
regulation of cell adhesion
|
0.0487785466817079
|
748
|
97
|
11
|
0.11340206185567
|
0.0147058823529412
|
|
GO:BP
|
GO:0001738
|
morphogenesis of a polarized epithelium
|
0.0487785466817079
|
152
|
106
|
5
|
0.0471698113207547
|
0.0328947368421053
|
|
GO:BP
|
GO:0007399
|
nervous system development
|
0.0493229359327963
|
2449
|
114
|
27
|
0.236842105263158
|
0.0110249081257656
|
|
GO:CC
|
GO:0005615
|
extracellular space
|
9.27055814531836e-06
|
3594
|
19
|
15
|
0.789473684210526
|
0.00417362270450751
|
|
GO:CC
|
GO:1903561
|
extracellular vesicle
|
3.3260187517566e-05
|
2261
|
14
|
10
|
0.714285714285714
|
0.00442282176028306
|
|
GO:CC
|
GO:0070062
|
extracellular exosome
|
3.3260187517566e-05
|
2176
|
14
|
10
|
0.714285714285714
|
0.00459558823529412
|
|
GO:CC
|
GO:0043230
|
extracellular organelle
|
3.3260187517566e-05
|
2263
|
14
|
10
|
0.714285714285714
|
0.00441891294741494
|
|
GO:CC
|
GO:0005576
|
extracellular region
|
5.19230830831883e-05
|
4567
|
21
|
16
|
0.761904761904762
|
0.00350339391285308
|
|
GO:CC
|
GO:0031982
|
vesicle
|
0.00126203237829676
|
4055
|
15
|
11
|
0.733333333333333
|
0.00271270036991369
|
|
GO:CC
|
GO:0031225
|
anchored component of membrane
|
0.0123556807986152
|
174
|
97
|
6
|
0.0618556701030928
|
0.0344827586206897
|
|
GO:CC
|
GO:0031012
|
extracellular matrix
|
0.0431962025316458
|
562
|
89
|
9
|
0.101123595505618
|
0.0160142348754448
|
|
GO:CC
|
GO:0030312
|
external encapsulating structure
|
0.0431962025316458
|
563
|
89
|
9
|
0.101123595505618
|
0.0159857904085258
|
|
GO:MF
|
GO:0070492
|
oligosaccharide binding
|
1.90186681258493e-07
|
15
|
3
|
3
|
1
|
0.2
|
|
GO:MF
|
GO:0042834
|
peptidoglycan binding
|
1.90186681258493e-07
|
18
|
3
|
3
|
1
|
0.166666666666667
|
|
GO:MF
|
GO:0005539
|
glycosaminoglycan binding
|
0.000344734491634493
|
238
|
3
|
3
|
1
|
0.0126050420168067
|
|
GO:MF
|
GO:0030246
|
carbohydrate binding
|
0.000399534787189738
|
275
|
3
|
3
|
1
|
0.0109090909090909
|
|
GO:MF
|
GO:0017147
|
Wnt-protein binding
|
0.0229253505115581
|
30
|
89
|
3
|
0.0337078651685393
|
0.1
|
|
GO:MF
|
GO:0046625
|
sphingolipid binding
|
0.0229253505115581
|
27
|
101
|
3
|
0.0297029702970297
|
0.111111111111111
|
|
GO:MF
|
GO:0060089
|
molecular transducer activity
|
0.0229253505115581
|
1458
|
3
|
3
|
1
|
0.00205761316872428
|
|
GO:MF
|
GO:0038023
|
signaling receptor activity
|
0.0229253505115581
|
1458
|
3
|
3
|
1
|
0.00205761316872428
|
|
GO:MF
|
GO:0097367
|
carbohydrate derivative binding
|
0.0374315957274293
|
2285
|
8
|
5
|
0.625
|
0.00218818380743982
|
|
REAC
|
REAC:R-HSA-6803157
|
Antimicrobial peptides
|
0.00277602751670245
|
80
|
18
|
4
|
0.222222222222222
|
0.05
|
|
REAC
|
REAC:R-HSA-168249
|
Innate Immune System
|
0.0421064354165665
|
1073
|
20
|
8
|
0.4
|
0.00745573159366263
|
|
REAC
|
REAC:R-HSA-977606
|
Regulation of Complement cascade
|
0.0421064354165665
|
101
|
17
|
3
|
0.176470588235294
|
0.0297029702970297
|
|
REAC
|
REAC:R-HSA-166658
|
Complement cascade
|
0.04439069115286
|
111
|
17
|
3
|
0.176470588235294
|
0.027027027027027
|