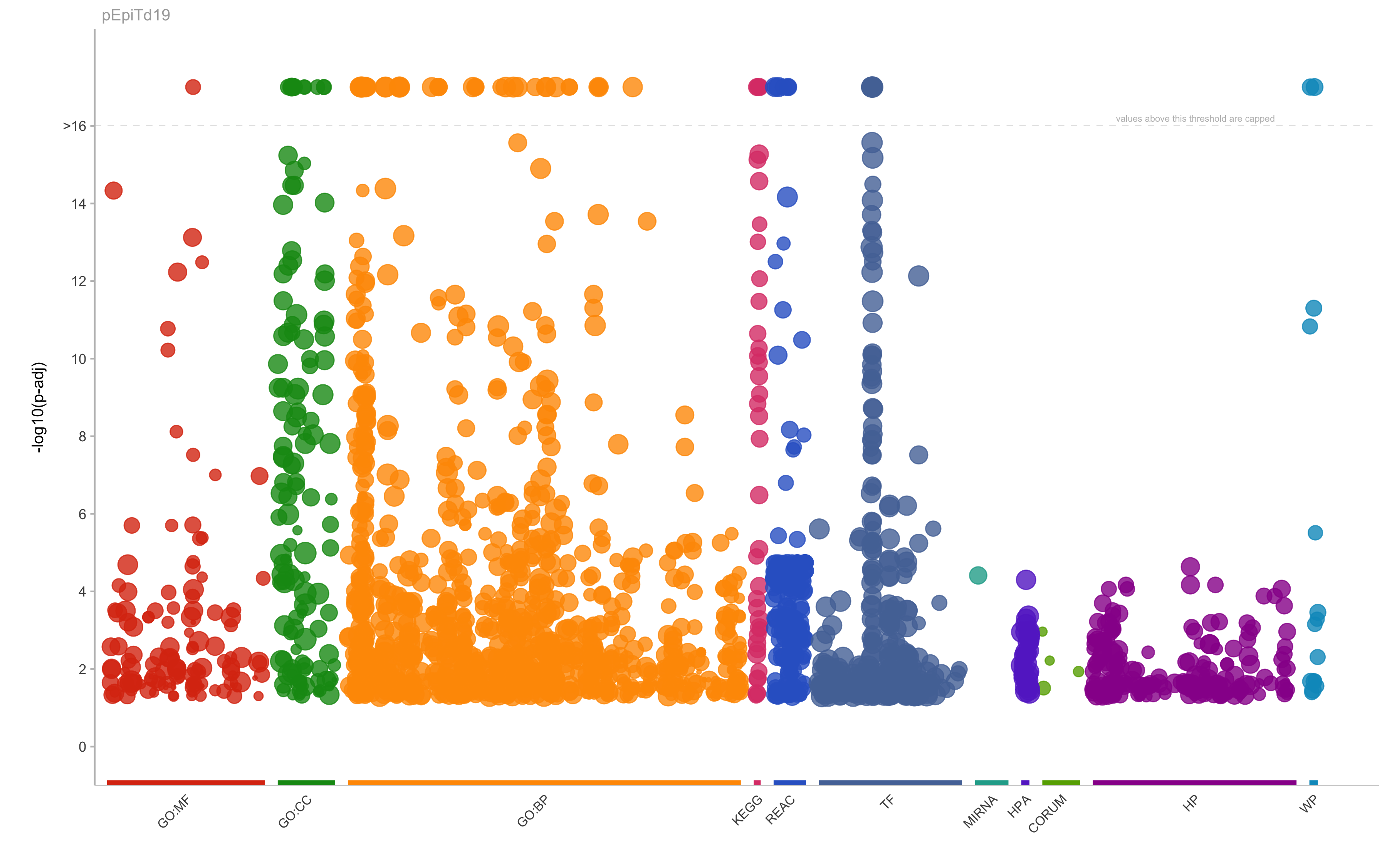
|
GO:BP
|
GO:0006955
|
immune response
|
3.66242830621905e-37
|
2311
|
123
|
79
|
0.642276422764228
|
0.0341843357853743
|
|
GO:BP
|
GO:0034341
|
response to interferon-gamma
|
4.59972952971057e-34
|
196
|
135
|
34
|
0.251851851851852
|
0.173469387755102
|
|
GO:BP
|
GO:0098542
|
defense response to other organism
|
8.21683957223981e-34
|
1218
|
135
|
62
|
0.459259259259259
|
0.0509031198686371
|
|
GO:BP
|
GO:0043207
|
response to external biotic stimulus
|
2.66870388289058e-32
|
1594
|
135
|
67
|
0.496296296296296
|
0.0420326223337516
|
|
GO:BP
|
GO:0051707
|
response to other organism
|
2.66870388289058e-32
|
1592
|
135
|
67
|
0.496296296296296
|
0.0420854271356784
|
|
GO:BP
|
GO:0009607
|
response to biotic stimulus
|
7.60882665213554e-32
|
1626
|
135
|
67
|
0.496296296296296
|
0.0412054120541205
|
|
GO:BP
|
GO:0002478
|
antigen processing and presentation of exogenous peptide antigen
|
1.00654575752321e-31
|
174
|
41
|
22
|
0.536585365853659
|
0.126436781609195
|
|
GO:BP
|
GO:0019882
|
antigen processing and presentation
|
1.47934913539614e-31
|
234
|
25
|
20
|
0.8
|
0.0854700854700855
|
|
GO:BP
|
GO:0045087
|
innate immune response
|
2.00211745368197e-31
|
998
|
135
|
55
|
0.407407407407407
|
0.0551102204408818
|
|
GO:BP
|
GO:0019884
|
antigen processing and presentation of exogenous antigen
|
2.00211745368197e-31
|
182
|
41
|
22
|
0.536585365853659
|
0.120879120879121
|
|
GO:BP
|
GO:0048002
|
antigen processing and presentation of peptide antigen
|
5.94992101703068e-31
|
192
|
25
|
19
|
0.76
|
0.0989583333333333
|
|
GO:BP
|
GO:0002376
|
immune system process
|
1.79269526373085e-30
|
3278
|
132
|
86
|
0.651515151515151
|
0.026235509456986
|
|
GO:BP
|
GO:0006952
|
defense response
|
1.99909695229012e-29
|
1818
|
138
|
68
|
0.492753623188406
|
0.0374037403740374
|
|
GO:BP
|
GO:0019221
|
cytokine-mediated signaling pathway
|
1.99909695229012e-29
|
810
|
135
|
49
|
0.362962962962963
|
0.0604938271604938
|
|
GO:BP
|
GO:0060333
|
interferon-gamma-mediated signaling pathway
|
2.09947467098898e-28
|
88
|
71
|
20
|
0.28169014084507
|
0.227272727272727
|
|
GO:BP
|
GO:0071346
|
cellular response to interferon-gamma
|
2.45667966298665e-27
|
175
|
135
|
28
|
0.207407407407407
|
0.16
|
|
GO:BP
|
GO:0034097
|
response to cytokine
|
4.30706493768819e-27
|
1226
|
135
|
55
|
0.407407407407407
|
0.0448613376835237
|
|
GO:BP
|
GO:0050776
|
regulation of immune response
|
9.2070926694886e-27
|
996
|
111
|
46
|
0.414414414414414
|
0.0461847389558233
|
|
GO:BP
|
GO:0044419
|
biological process involved in interspecies interaction between organisms
|
2.06654712141171e-26
|
2255
|
135
|
70
|
0.518518518518518
|
0.0310421286031042
|
|
GO:BP
|
GO:0071345
|
cellular response to cytokine stimulus
|
2.12471954271528e-26
|
1132
|
71
|
39
|
0.549295774647887
|
0.034452296819788
|
|
GO:BP
|
GO:0002682
|
regulation of immune system process
|
8.54491697663114e-25
|
1617
|
135
|
59
|
0.437037037037037
|
0.0364873222016079
|
|
GO:BP
|
GO:0050778
|
positive regulation of immune response
|
3.79869049627796e-23
|
753
|
109
|
38
|
0.348623853211009
|
0.050464807436919
|
|
GO:BP
|
GO:0002684
|
positive regulation of immune system process
|
7.67475245173168e-23
|
1065
|
109
|
43
|
0.394495412844037
|
0.0403755868544601
|
|
GO:BP
|
GO:0019883
|
antigen processing and presentation of endogenous antigen
|
3.54718722025523e-22
|
26
|
36
|
11
|
0.305555555555556
|
0.423076923076923
|
|
GO:BP
|
GO:0002483
|
antigen processing and presentation of endogenous peptide antigen
|
2.85614261322658e-21
|
19
|
36
|
10
|
0.277777777777778
|
0.526315789473684
|
|
GO:BP
|
GO:0009605
|
response to external stimulus
|
4.23333159457611e-21
|
2944
|
135
|
72
|
0.533333333333333
|
0.0244565217391304
|
|
GO:BP
|
GO:0002479
|
antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent
|
4.60513237479235e-20
|
75
|
40
|
13
|
0.325
|
0.173333333333333
|
|
GO:BP
|
GO:0002250
|
adaptive immune response
|
7.15659969139101e-20
|
689
|
111
|
34
|
0.306306306306306
|
0.0493468795355588
|
|
GO:BP
|
GO:0042590
|
antigen processing and presentation of exogenous peptide antigen via MHC class I
|
1.05987201968638e-19
|
80
|
40
|
13
|
0.325
|
0.1625
|
|
GO:BP
|
GO:0002252
|
immune effector process
|
1.23713278070741e-19
|
1309
|
84
|
38
|
0.452380952380952
|
0.0290297937356761
|
|
GO:BP
|
GO:0060337
|
type I interferon signaling pathway
|
1.49194208053034e-19
|
95
|
79
|
16
|
0.20253164556962
|
0.168421052631579
|
|
GO:BP
|
GO:0071357
|
cellular response to type I interferon
|
1.72868302429129e-19
|
96
|
79
|
16
|
0.20253164556962
|
0.166666666666667
|
|
GO:BP
|
GO:0034340
|
response to type I interferon
|
3.98182299202798e-19
|
101
|
79
|
16
|
0.20253164556962
|
0.158415841584158
|
|
GO:BP
|
GO:0002474
|
antigen processing and presentation of peptide antigen via MHC class I
|
1.72831212410566e-18
|
99
|
40
|
13
|
0.325
|
0.131313131313131
|
|
GO:BP
|
GO:0002831
|
regulation of response to biotic stimulus
|
3.41273513910309e-17
|
415
|
135
|
28
|
0.207407407407407
|
0.0674698795180723
|
|
GO:BP
|
GO:0009615
|
response to virus
|
7.94621639473032e-17
|
364
|
131
|
26
|
0.198473282442748
|
0.0714285714285714
|
|
GO:BP
|
GO:0045088
|
regulation of innate immune response
|
2.73052757553718e-16
|
309
|
69
|
19
|
0.27536231884058
|
0.0614886731391586
|
|
GO:BP
|
GO:0048584
|
positive regulation of response to stimulus
|
1.25381626426259e-15
|
2344
|
111
|
51
|
0.459459459459459
|
0.0217576791808874
|
|
GO:BP
|
GO:0006950
|
response to stress
|
4.13230413047097e-15
|
4170
|
132
|
75
|
0.568181818181818
|
0.0179856115107914
|
|
GO:BP
|
GO:0002480
|
antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent
|
4.63225296927719e-15
|
8
|
23
|
6
|
0.260869565217391
|
0.75
|
|
GO:BP
|
GO:0071310
|
cellular response to organic substance
|
1.93096922454799e-14
|
2776
|
71
|
41
|
0.577464788732394
|
0.0147694524495677
|
|
GO:BP
|
GO:0051607
|
defense response to virus
|
2.88301565029884e-14
|
267
|
131
|
21
|
0.16030534351145
|
0.0786516853932584
|
|
GO:BP
|
GO:0140546
|
defense response to symbiont
|
2.88301565029884e-14
|
267
|
131
|
21
|
0.16030534351145
|
0.0786516853932584
|
|
GO:BP
|
GO:0010033
|
response to organic substance
|
6.77595420818685e-14
|
3412
|
58
|
39
|
0.672413793103448
|
0.0114302461899179
|
|
GO:BP
|
GO:0001916
|
positive regulation of T cell mediated cytotoxicity
|
8.96185049487488e-14
|
26
|
23
|
7
|
0.304347826086957
|
0.269230769230769
|
|
GO:BP
|
GO:0050852
|
T cell receptor signaling pathway
|
1.10518776790813e-13
|
203
|
57
|
14
|
0.245614035087719
|
0.0689655172413793
|
|
GO:BP
|
GO:0002504
|
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II
|
2.33053206401463e-13
|
103
|
90
|
13
|
0.144444444444444
|
0.12621359223301
|
|
GO:BP
|
GO:0002253
|
activation of immune response
|
4.19005139351252e-13
|
568
|
92
|
23
|
0.25
|
0.0404929577464789
|
|
GO:BP
|
GO:0007166
|
cell surface receptor signaling pathway
|
6.88448018496279e-13
|
3210
|
107
|
54
|
0.504672897196262
|
0.0168224299065421
|
|
GO:BP
|
GO:0001914
|
regulation of T cell mediated cytotoxicity
|
8.18808420865267e-13
|
35
|
23
|
7
|
0.304347826086957
|
0.2
|
|
GO:BP
|
GO:0002697
|
regulation of immune effector process
|
9.99743716256107e-13
|
461
|
116
|
23
|
0.198275862068966
|
0.0498915401301518
|
|
GO:BP
|
GO:0002683
|
negative regulation of immune system process
|
1.14680911333791e-12
|
420
|
91
|
20
|
0.21978021978022
|
0.0476190476190476
|
|
GO:BP
|
GO:0001816
|
cytokine production
|
2.1914045731818e-12
|
844
|
57
|
21
|
0.368421052631579
|
0.0248815165876777
|
|
GO:BP
|
GO:0070661
|
leukocyte proliferation
|
2.1914045731818e-12
|
325
|
135
|
21
|
0.155555555555556
|
0.0646153846153846
|
|
GO:BP
|
GO:0031347
|
regulation of defense response
|
2.24194542000038e-12
|
704
|
135
|
29
|
0.214814814814815
|
0.0411931818181818
|
|
GO:BP
|
GO:0019886
|
antigen processing and presentation of exogenous peptide antigen via MHC class II
|
2.70725685495183e-12
|
96
|
90
|
12
|
0.133333333333333
|
0.125
|
|
GO:BP
|
GO:0001913
|
T cell mediated cytotoxicity
|
2.86600845824797e-12
|
42
|
23
|
7
|
0.304347826086957
|
0.166666666666667
|
|
GO:BP
|
GO:0019885
|
antigen processing and presentation of endogenous peptide antigen via MHC class I
|
3.74638295517425e-12
|
13
|
36
|
6
|
0.166666666666667
|
0.461538461538462
|
|
GO:BP
|
GO:0002495
|
antigen processing and presentation of peptide antigen via MHC class II
|
4.25246767743089e-12
|
100
|
90
|
12
|
0.133333333333333
|
0.12
|
|
GO:BP
|
GO:0070663
|
regulation of leukocyte proliferation
|
4.95645962357047e-12
|
251
|
123
|
18
|
0.146341463414634
|
0.0717131474103586
|
|
GO:BP
|
GO:0046651
|
lymphocyte proliferation
|
6.05965982843096e-12
|
294
|
107
|
18
|
0.168224299065421
|
0.0612244897959184
|
|
GO:BP
|
GO:0032943
|
mononuclear cell proliferation
|
7.04414215840667e-12
|
297
|
107
|
18
|
0.168224299065421
|
0.0606060606060606
|
|
GO:BP
|
GO:0002711
|
positive regulation of T cell mediated immunity
|
7.04414215840667e-12
|
48
|
23
|
7
|
0.304347826086957
|
0.145833333333333
|
|
GO:BP
|
GO:0032101
|
regulation of response to external stimulus
|
8.21646277879746e-12
|
1051
|
56
|
22
|
0.392857142857143
|
0.0209324452901998
|
|
GO:BP
|
GO:0001817
|
regulation of cytokine production
|
9.25162936103185e-12
|
806
|
57
|
20
|
0.350877192982456
|
0.0248138957816377
|
|
GO:BP
|
GO:0001912
|
positive regulation of leukocyte mediated cytotoxicity
|
1.05482743716545e-11
|
51
|
23
|
7
|
0.304347826086957
|
0.137254901960784
|
|
GO:BP
|
GO:0050670
|
regulation of lymphocyte proliferation
|
1.39055821113867e-11
|
230
|
123
|
17
|
0.138211382113821
|
0.0739130434782609
|
|
GO:BP
|
GO:0070887
|
cellular response to chemical stimulus
|
1.39814907088316e-11
|
3393
|
71
|
41
|
0.577464788732394
|
0.0120837017388742
|
|
GO:BP
|
GO:0042221
|
response to chemical
|
1.45046909007063e-11
|
4781
|
58
|
42
|
0.724137931034483
|
0.00878477306002928
|
|
GO:BP
|
GO:0032944
|
regulation of mononuclear cell proliferation
|
1.53306654440232e-11
|
232
|
123
|
17
|
0.138211382113821
|
0.0732758620689655
|
|
GO:BP
|
GO:0016032
|
viral process
|
2.15849349839303e-11
|
942
|
27
|
15
|
0.555555555555556
|
0.0159235668789809
|
|
GO:BP
|
GO:0050851
|
antigen receptor-mediated signaling pathway
|
2.30033305358567e-11
|
323
|
92
|
17
|
0.184782608695652
|
0.0526315789473684
|
|
GO:BP
|
GO:0031343
|
positive regulation of cell killing
|
2.79280361798265e-11
|
59
|
23
|
7
|
0.304347826086957
|
0.11864406779661
|
|
GO:BP
|
GO:0042098
|
T cell proliferation
|
2.85363382126778e-11
|
205
|
123
|
16
|
0.130081300813008
|
0.0780487804878049
|
|
GO:BP
|
GO:0002460
|
adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
|
3.17524837064523e-11
|
363
|
111
|
19
|
0.171171171171171
|
0.0523415977961433
|
|
GO:BP
|
GO:0044403
|
biological process involved in symbiotic interaction
|
4.85854831064754e-11
|
1001
|
27
|
15
|
0.555555555555556
|
0.014985014985015
|
|
GO:BP
|
GO:0002501
|
peptide antigen assembly with MHC protein complex
|
8.41372120512441e-11
|
6
|
60
|
5
|
0.0833333333333333
|
0.833333333333333
|
|
GO:BP
|
GO:0002503
|
peptide antigen assembly with MHC class II protein complex
|
1.13536634932776e-10
|
4
|
25
|
4
|
0.16
|
1
|
|
GO:BP
|
GO:0001775
|
cell activation
|
1.13536634932776e-10
|
1495
|
43
|
21
|
0.488372093023256
|
0.0140468227424749
|
|
GO:BP
|
GO:0002399
|
MHC class II protein complex assembly
|
1.13536634932776e-10
|
4
|
25
|
4
|
0.16
|
1
|
|
GO:BP
|
GO:0045321
|
leukocyte activation
|
1.21315291547102e-10
|
1332
|
52
|
22
|
0.423076923076923
|
0.0165165165165165
|
|
GO:BP
|
GO:0045824
|
negative regulation of innate immune response
|
1.21315291547102e-10
|
66
|
69
|
9
|
0.130434782608696
|
0.136363636363636
|
|
GO:BP
|
GO:0001910
|
regulation of leukocyte mediated cytotoxicity
|
1.26549767274203e-10
|
74
|
23
|
7
|
0.304347826086957
|
0.0945945945945946
|
|
GO:BP
|
GO:0002709
|
regulation of T cell mediated immunity
|
1.26549767274203e-10
|
74
|
23
|
7
|
0.304347826086957
|
0.0945945945945946
|
|
GO:BP
|
GO:0002449
|
lymphocyte mediated immunity
|
1.94086140752232e-10
|
354
|
111
|
18
|
0.162162162162162
|
0.0508474576271186
|
|
GO:BP
|
GO:0002699
|
positive regulation of immune effector process
|
2.56297453068392e-10
|
215
|
25
|
9
|
0.36
|
0.0418604651162791
|
|
GO:BP
|
GO:0002396
|
MHC protein complex assembly
|
2.59972412768204e-10
|
7
|
60
|
5
|
0.0833333333333333
|
0.714285714285714
|
|
GO:BP
|
GO:0050896
|
response to stimulus
|
3.70085053964737e-10
|
9407
|
132
|
106
|
0.803030303030303
|
0.0112682045285426
|
|
GO:BP
|
GO:0048583
|
regulation of response to stimulus
|
5.02220830324565e-10
|
4278
|
58
|
38
|
0.655172413793103
|
0.00888265544647031
|
|
GO:BP
|
GO:0042129
|
regulation of T cell proliferation
|
5.45397029006437e-10
|
176
|
123
|
14
|
0.113821138211382
|
0.0795454545454545
|
|
GO:BP
|
GO:0031341
|
regulation of cell killing
|
6.02648031583948e-10
|
93
|
23
|
7
|
0.304347826086957
|
0.0752688172043011
|
|
GO:BP
|
GO:0050777
|
negative regulation of immune response
|
6.02648031583948e-10
|
157
|
69
|
11
|
0.159420289855072
|
0.0700636942675159
|
|
GO:BP
|
GO:0042110
|
T cell activation
|
6.41199312899383e-10
|
489
|
57
|
15
|
0.263157894736842
|
0.0306748466257669
|
|
GO:BP
|
GO:0002824
|
positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
|
7.36340390744094e-10
|
96
|
23
|
7
|
0.304347826086957
|
0.0729166666666667
|
|
GO:BP
|
GO:0002456
|
T cell mediated immunity
|
8.44350649035056e-10
|
98
|
23
|
7
|
0.304347826086957
|
0.0714285714285714
|
|
GO:BP
|
GO:0002429
|
immune response-activating cell surface receptor signaling pathway
|
8.61397960801188e-10
|
477
|
92
|
18
|
0.195652173913043
|
0.0377358490566038
|
|
GO:BP
|
GO:0002757
|
immune response-activating signal transduction
|
8.61397960801188e-10
|
477
|
92
|
18
|
0.195652173913043
|
0.0377358490566038
|
|
GO:BP
|
GO:0032103
|
positive regulation of response to external stimulus
|
8.61397960801188e-10
|
513
|
86
|
18
|
0.209302325581395
|
0.0350877192982456
|
|
GO:BP
|
GO:0002819
|
regulation of adaptive immune response
|
9.69114724084812e-10
|
167
|
111
|
13
|
0.117117117117117
|
0.0778443113772455
|
|
GO:BP
|
GO:0002821
|
positive regulation of adaptive immune response
|
9.9493196745134e-10
|
101
|
23
|
7
|
0.304347826086957
|
0.0693069306930693
|
|
GO:BP
|
GO:0046649
|
lymphocyte activation
|
1.12606888721385e-09
|
771
|
111
|
24
|
0.216216216216216
|
0.0311284046692607
|
|
GO:BP
|
GO:0002708
|
positive regulation of lymphocyte mediated immunity
|
1.20194962035185e-09
|
104
|
23
|
7
|
0.304347826086957
|
0.0673076923076923
|
|
GO:BP
|
GO:0051249
|
regulation of lymphocyte activation
|
1.31770171653115e-09
|
518
|
123
|
21
|
0.170731707317073
|
0.0405405405405405
|
|
GO:BP
|
GO:0070665
|
positive regulation of leukocyte proliferation
|
1.34052153531879e-09
|
155
|
123
|
13
|
0.105691056910569
|
0.0838709677419355
|
|
GO:BP
|
GO:0001909
|
leukocyte mediated cytotoxicity
|
1.42989315081153e-09
|
107
|
23
|
7
|
0.304347826086957
|
0.0654205607476635
|
|
GO:BP
|
GO:0002768
|
immune response-regulating cell surface receptor signaling pathway
|
2.53662742321135e-09
|
512
|
92
|
18
|
0.195652173913043
|
0.03515625
|
|
GO:BP
|
GO:0002764
|
immune response-regulating signaling pathway
|
2.76352663961724e-09
|
515
|
92
|
18
|
0.195652173913043
|
0.0349514563106796
|
|
GO:BP
|
GO:0050865
|
regulation of cell activation
|
2.76600848012326e-09
|
655
|
135
|
24
|
0.177777777777778
|
0.0366412213740458
|
|
GO:BP
|
GO:0050863
|
regulation of T cell activation
|
2.77193002982157e-09
|
337
|
30
|
10
|
0.333333333333333
|
0.029673590504451
|
|
GO:BP
|
GO:1903037
|
regulation of leukocyte cell-cell adhesion
|
2.827590785539e-09
|
338
|
30
|
10
|
0.333333333333333
|
0.029585798816568
|
|
GO:BP
|
GO:0002703
|
regulation of leukocyte mediated immunity
|
2.88380828635302e-09
|
205
|
23
|
8
|
0.347826086956522
|
0.0390243902439024
|
|
GO:BP
|
GO:0002705
|
positive regulation of leukocyte mediated immunity
|
3.40365311866266e-09
|
122
|
23
|
7
|
0.304347826086957
|
0.0573770491803279
|
|
GO:BP
|
GO:0002832
|
negative regulation of response to biotic stimulus
|
4.04943384934485e-09
|
100
|
69
|
9
|
0.130434782608696
|
0.09
|
|
GO:BP
|
GO:0002833
|
positive regulation of response to biotic stimulus
|
4.16539136159435e-09
|
250
|
85
|
13
|
0.152941176470588
|
0.052
|
|
GO:BP
|
GO:0007165
|
signal transduction
|
5.41226972182121e-09
|
6309
|
69
|
50
|
0.72463768115942
|
0.00792518624187668
|
|
GO:BP
|
GO:0002428
|
antigen processing and presentation of peptide antigen via MHC class Ib
|
5.45371488455434e-09
|
6
|
36
|
4
|
0.111111111111111
|
0.666666666666667
|
|
GO:BP
|
GO:0050671
|
positive regulation of lymphocyte proliferation
|
5.8296987472133e-09
|
141
|
123
|
12
|
0.0975609756097561
|
0.0851063829787234
|
|
GO:BP
|
GO:0045953
|
negative regulation of natural killer cell mediated cytotoxicity
|
6.1254262391403e-09
|
16
|
42
|
5
|
0.119047619047619
|
0.3125
|
|
GO:BP
|
GO:0032946
|
positive regulation of mononuclear cell proliferation
|
6.22529996777478e-09
|
142
|
123
|
12
|
0.0975609756097561
|
0.0845070422535211
|
|
GO:BP
|
GO:0007159
|
leukocyte cell-cell adhesion
|
7.14827587544798e-09
|
375
|
30
|
10
|
0.333333333333333
|
0.0266666666666667
|
|
GO:BP
|
GO:0002716
|
negative regulation of natural killer cell mediated immunity
|
8.44813177183043e-09
|
17
|
42
|
5
|
0.119047619047619
|
0.294117647058824
|
|
GO:BP
|
GO:0050870
|
positive regulation of T cell activation
|
9.60221267074635e-09
|
219
|
25
|
8
|
0.32
|
0.0365296803652968
|
|
GO:BP
|
GO:0002468
|
dendritic cell antigen processing and presentation
|
9.62246021122425e-09
|
14
|
3
|
3
|
1
|
0.214285714285714
|
|
GO:BP
|
GO:0045089
|
positive regulation of innate immune response
|
9.6697390691042e-09
|
216
|
85
|
12
|
0.141176470588235
|
0.0555555555555556
|
|
GO:BP
|
GO:0002520
|
immune system development
|
9.73066495332377e-09
|
1020
|
42
|
16
|
0.380952380952381
|
0.0156862745098039
|
|
GO:BP
|
GO:0001819
|
positive regulation of cytokine production
|
1.06068258909222e-08
|
452
|
53
|
13
|
0.245283018867925
|
0.0287610619469027
|
|
GO:BP
|
GO:0002477
|
antigen processing and presentation of exogenous peptide antigen via MHC class Ib
|
1.16492382478404e-08
|
3
|
15
|
3
|
0.2
|
1
|
|
GO:BP
|
GO:0002706
|
regulation of lymphocyte mediated immunity
|
1.34067092746636e-08
|
151
|
23
|
7
|
0.304347826086957
|
0.0463576158940397
|
|
GO:BP
|
GO:0002822
|
regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
|
1.39341471914187e-08
|
152
|
23
|
7
|
0.304347826086957
|
0.0460526315789474
|
|
GO:BP
|
GO:0080134
|
regulation of response to stress
|
1.61172949102452e-08
|
1443
|
56
|
21
|
0.375
|
0.0145530145530146
|
|
GO:BP
|
GO:0051251
|
positive regulation of lymphocyte activation
|
1.87389259132035e-08
|
365
|
25
|
9
|
0.36
|
0.0246575342465753
|
|
GO:BP
|
GO:1903039
|
positive regulation of leukocyte cell-cell adhesion
|
1.89616892669688e-08
|
241
|
25
|
8
|
0.32
|
0.033195020746888
|
|
GO:BP
|
GO:0002694
|
regulation of leukocyte activation
|
1.90158118006967e-08
|
610
|
111
|
20
|
0.18018018018018
|
0.0327868852459016
|
|
GO:BP
|
GO:0001911
|
negative regulation of leukocyte mediated cytotoxicity
|
1.90158118006967e-08
|
20
|
42
|
5
|
0.119047619047619
|
0.25
|
|
GO:BP
|
GO:0002718
|
regulation of cytokine production involved in immune response
|
2.01485261290545e-08
|
85
|
23
|
6
|
0.260869565217391
|
0.0705882352941176
|
|
GO:BP
|
GO:0022407
|
regulation of cell-cell adhesion
|
3.31434536414629e-08
|
445
|
30
|
10
|
0.333333333333333
|
0.0224719101123595
|
|
GO:BP
|
GO:0001906
|
cell killing
|
3.52082104394486e-08
|
175
|
23
|
7
|
0.304347826086957
|
0.04
|
|
GO:BP
|
GO:0002367
|
cytokine production involved in immune response
|
3.64293235720454e-08
|
94
|
23
|
6
|
0.260869565217391
|
0.0638297872340425
|
|
GO:BP
|
GO:0002486
|
antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent
|
4.08403354218613e-08
|
3
|
23
|
3
|
0.130434782608696
|
1
|
|
GO:BP
|
GO:0002704
|
negative regulation of leukocyte mediated immunity
|
4.08403354218613e-08
|
51
|
42
|
6
|
0.142857142857143
|
0.117647058823529
|
|
GO:BP
|
GO:0002484
|
antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway
|
4.08403354218613e-08
|
3
|
23
|
3
|
0.130434782608696
|
1
|
|
GO:BP
|
GO:0031342
|
negative regulation of cell killing
|
4.88604660886703e-08
|
24
|
42
|
5
|
0.119047619047619
|
0.208333333333333
|
|
GO:BP
|
GO:0002696
|
positive regulation of leukocyte activation
|
4.97253698253575e-08
|
412
|
25
|
9
|
0.36
|
0.0218446601941748
|
|
GO:BP
|
GO:0002440
|
production of molecular mediator of immune response
|
5.73048010263686e-08
|
189
|
23
|
7
|
0.304347826086957
|
0.037037037037037
|
|
GO:BP
|
GO:0022409
|
positive regulation of cell-cell adhesion
|
6.26108604854969e-08
|
284
|
25
|
8
|
0.32
|
0.028169014084507
|
|
GO:BP
|
GO:0050867
|
positive regulation of cell activation
|
6.26108604854969e-08
|
424
|
25
|
9
|
0.36
|
0.0212264150943396
|
|
GO:BP
|
GO:0002443
|
leukocyte mediated immunity
|
6.41891932694056e-08
|
877
|
47
|
15
|
0.319148936170213
|
0.0171037628278221
|
|
GO:BP
|
GO:0034612
|
response to tumor necrosis factor
|
7.56276144514312e-08
|
331
|
135
|
16
|
0.118518518518519
|
0.0483383685800604
|
|
GO:BP
|
GO:0023052
|
signaling
|
8.74454679663116e-08
|
6810
|
69
|
50
|
0.72463768115942
|
0.00734214390602056
|
|
GO:BP
|
GO:0007154
|
cell communication
|
9.68717356822792e-08
|
6829
|
69
|
50
|
0.72463768115942
|
0.00732171621027969
|
|
GO:BP
|
GO:0009617
|
response to bacterium
|
1.30601068772348e-07
|
743
|
135
|
23
|
0.17037037037037
|
0.0309555854643338
|
|
GO:BP
|
GO:0048585
|
negative regulation of response to stimulus
|
1.32207343908919e-07
|
1733
|
58
|
22
|
0.379310344827586
|
0.0126947489901904
|
|
GO:BP
|
GO:0002720
|
positive regulation of cytokine production involved in immune response
|
1.32369043788347e-07
|
54
|
23
|
5
|
0.217391304347826
|
0.0925925925925926
|
|
GO:BP
|
GO:0070555
|
response to interleukin-1
|
1.65090031622434e-07
|
215
|
135
|
13
|
0.0962962962962963
|
0.0604651162790698
|
|
GO:BP
|
GO:0071356
|
cellular response to tumor necrosis factor
|
1.90781845718078e-07
|
306
|
135
|
15
|
0.111111111111111
|
0.0490196078431373
|
|
GO:BP
|
GO:0002475
|
antigen processing and presentation via MHC class Ib
|
1.91399665926554e-07
|
13
|
36
|
4
|
0.111111111111111
|
0.307692307692308
|
|
GO:BP
|
GO:0031349
|
positive regulation of defense response
|
2.18245650623649e-07
|
362
|
69
|
12
|
0.173913043478261
|
0.0331491712707182
|
|
GO:BP
|
GO:0030097
|
hemopoiesis
|
2.18245650623649e-07
|
928
|
42
|
14
|
0.333333333333333
|
0.0150862068965517
|
|
GO:BP
|
GO:0046596
|
regulation of viral entry into host cell
|
2.25759007100174e-07
|
43
|
67
|
6
|
0.0895522388059701
|
0.13953488372093
|
|
GO:BP
|
GO:1903900
|
regulation of viral life cycle
|
2.90807313939651e-07
|
153
|
131
|
11
|
0.083969465648855
|
0.0718954248366013
|
|
GO:BP
|
GO:0050792
|
regulation of viral process
|
3.00766856821035e-07
|
192
|
131
|
12
|
0.0916030534351145
|
0.0625
|
|
GO:BP
|
GO:0046634
|
regulation of alpha-beta T cell activation
|
3.14689012470484e-07
|
102
|
52
|
7
|
0.134615384615385
|
0.0686274509803922
|
|
GO:BP
|
GO:0048534
|
hematopoietic or lymphoid organ development
|
3.51755782776239e-07
|
966
|
42
|
14
|
0.333333333333333
|
0.0144927536231884
|
|
GO:BP
|
GO:0042127
|
regulation of cell population proliferation
|
3.52705083360832e-07
|
1774
|
55
|
21
|
0.381818181818182
|
0.0118376550169109
|
|
GO:BP
|
GO:0008283
|
cell population proliferation
|
3.52705083360832e-07
|
2054
|
57
|
23
|
0.403508771929825
|
0.0111976630963973
|
|
GO:BP
|
GO:0035456
|
response to interferon-beta
|
4.56533955502966e-07
|
30
|
53
|
5
|
0.0943396226415094
|
0.166666666666667
|
|
GO:BP
|
GO:0002700
|
regulation of production of molecular mediator of immune response
|
4.60707587089492e-07
|
148
|
23
|
6
|
0.260869565217391
|
0.0405405405405405
|
|
GO:BP
|
GO:0043123
|
positive regulation of I-kappaB kinase/NF-kappaB signaling
|
4.83547523167463e-07
|
185
|
67
|
9
|
0.134328358208955
|
0.0486486486486487
|
|
GO:BP
|
GO:0043122
|
regulation of I-kappaB kinase/NF-kappaB signaling
|
5.09068717898725e-07
|
251
|
67
|
10
|
0.149253731343284
|
0.0398406374501992
|
|
GO:BP
|
GO:0043903
|
regulation of biological process involved in symbiotic interaction
|
5.23778326736017e-07
|
203
|
131
|
12
|
0.0916030534351145
|
0.0591133004926108
|
|
GO:BP
|
GO:0052372
|
modulation by symbiont of entry into host
|
5.30001554920287e-07
|
50
|
67
|
6
|
0.0895522388059701
|
0.12
|
|
GO:BP
|
GO:0031348
|
negative regulation of defense response
|
5.53645093062841e-07
|
246
|
69
|
10
|
0.144927536231884
|
0.040650406504065
|
|
GO:BP
|
GO:0002698
|
negative regulation of immune effector process
|
5.78693683900602e-07
|
125
|
47
|
7
|
0.148936170212766
|
0.056
|
|
GO:BP
|
GO:0042270
|
protection from natural killer cell mediated cytotoxicity
|
6.52749281829599e-07
|
6
|
23
|
3
|
0.130434782608696
|
0.5
|
|
GO:BP
|
GO:0042102
|
positive regulation of T cell proliferation
|
6.63505127462934e-07
|
104
|
123
|
9
|
0.0731707317073171
|
0.0865384615384615
|
|
GO:BP
|
GO:0042269
|
regulation of natural killer cell mediated cytotoxicity
|
6.6762092961888e-07
|
41
|
42
|
5
|
0.119047619047619
|
0.121951219512195
|
|
GO:BP
|
GO:0033209
|
tumor necrosis factor-mediated signaling pathway
|
7.76869522825617e-07
|
177
|
53
|
8
|
0.150943396226415
|
0.0451977401129944
|
|
GO:BP
|
GO:0051716
|
cellular response to stimulus
|
7.7797363871794e-07
|
7781
|
69
|
52
|
0.753623188405797
|
0.00668294563680761
|
|
GO:BP
|
GO:0030155
|
regulation of cell adhesion
|
8.27911141098362e-07
|
748
|
33
|
11
|
0.333333333333333
|
0.0147058823529412
|
|
GO:BP
|
GO:0002707
|
negative regulation of lymphocyte mediated immunity
|
8.3585541857074e-07
|
43
|
42
|
5
|
0.119047619047619
|
0.116279069767442
|
|
GO:BP
|
GO:0002366
|
leukocyte activation involved in immune response
|
8.46027592709853e-07
|
717
|
50
|
13
|
0.26
|
0.0181311018131102
|
|
GO:BP
|
GO:0046640
|
regulation of alpha-beta T cell proliferation
|
8.51705291688879e-07
|
35
|
52
|
5
|
0.0961538461538462
|
0.142857142857143
|
|
GO:BP
|
GO:0002263
|
cell activation involved in immune response
|
8.93184722977302e-07
|
721
|
50
|
13
|
0.26
|
0.0180305131761442
|
|
GO:BP
|
GO:0002715
|
regulation of natural killer cell mediated immunity
|
1.03454737748585e-06
|
45
|
42
|
5
|
0.119047619047619
|
0.111111111111111
|
|
GO:BP
|
GO:0032609
|
interferon-gamma production
|
1.21047559767654e-06
|
115
|
57
|
7
|
0.12280701754386
|
0.0608695652173913
|
|
GO:BP
|
GO:0002476
|
antigen processing and presentation of endogenous peptide antigen via MHC class Ib
|
1.23152331216575e-06
|
5
|
36
|
3
|
0.0833333333333333
|
0.6
|
|
GO:BP
|
GO:0046633
|
alpha-beta T cell proliferation
|
1.27376234110278e-06
|
38
|
52
|
5
|
0.0961538461538462
|
0.131578947368421
|
|
GO:BP
|
GO:0030593
|
neutrophil chemotaxis
|
1.27495154167182e-06
|
103
|
135
|
9
|
0.0666666666666667
|
0.087378640776699
|
|
GO:BP
|
GO:0045785
|
positive regulation of cell adhesion
|
1.36820112253336e-06
|
437
|
25
|
8
|
0.32
|
0.0183066361556064
|
|
GO:BP
|
GO:0051239
|
regulation of multicellular organismal process
|
1.71842833998112e-06
|
2861
|
57
|
26
|
0.456140350877193
|
0.00908773156239077
|
|
GO:BP
|
GO:0007249
|
I-kappaB kinase/NF-kappaB signaling
|
1.80824381425943e-06
|
291
|
67
|
10
|
0.149253731343284
|
0.0343642611683849
|
|
GO:BP
|
GO:0032831
|
positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation
|
1.9080663188719e-06
|
5
|
42
|
3
|
0.0714285714285714
|
0.6
|
|
GO:BP
|
GO:0032829
|
regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation
|
1.9080663188719e-06
|
5
|
42
|
3
|
0.0714285714285714
|
0.6
|
|
GO:BP
|
GO:0052548
|
regulation of endopeptidase activity
|
1.91372242459896e-06
|
437
|
58
|
11
|
0.189655172413793
|
0.0251716247139588
|
|
GO:BP
|
GO:0002230
|
positive regulation of defense response to virus by host
|
1.96586906395594e-06
|
33
|
131
|
6
|
0.0458015267175573
|
0.181818181818182
|
|
GO:BP
|
GO:0045582
|
positive regulation of T cell differentiation
|
1.99502030174758e-06
|
91
|
3
|
3
|
1
|
0.032967032967033
|
|
GO:BP
|
GO:0071347
|
cellular response to interleukin-1
|
2.2575841318806e-06
|
185
|
135
|
11
|
0.0814814814814815
|
0.0594594594594595
|
|
GO:BP
|
GO:0002702
|
positive regulation of production of molecular mediator of immune response
|
2.32038116555313e-06
|
100
|
23
|
5
|
0.217391304347826
|
0.05
|
|
GO:BP
|
GO:0045621
|
positive regulation of lymphocyte differentiation
|
2.94586797878038e-06
|
104
|
3
|
3
|
1
|
0.0288461538461538
|
|
GO:BP
|
GO:0035740
|
CD8-positive, alpha-beta T cell proliferation
|
3.25995519374363e-06
|
8
|
29
|
3
|
0.103448275862069
|
0.375
|
|
GO:BP
|
GO:2000564
|
regulation of CD8-positive, alpha-beta T cell proliferation
|
3.25995519374363e-06
|
8
|
29
|
3
|
0.103448275862069
|
0.375
|
|
GO:BP
|
GO:0052547
|
regulation of peptidase activity
|
3.57906396236891e-06
|
467
|
58
|
11
|
0.189655172413793
|
0.0235546038543897
|
|
GO:BP
|
GO:0002286
|
T cell activation involved in immune response
|
3.61508428243224e-06
|
112
|
3
|
3
|
1
|
0.0267857142857143
|
|
GO:BP
|
GO:0006959
|
humoral immune response
|
4.03668871659665e-06
|
384
|
86
|
12
|
0.13953488372093
|
0.03125
|
|
GO:BP
|
GO:0019058
|
viral life cycle
|
4.27781254022813e-06
|
353
|
131
|
14
|
0.106870229007634
|
0.0396600566572238
|
|
GO:BP
|
GO:0006521
|
regulation of cellular amino acid metabolic process
|
4.33562876206046e-06
|
64
|
40
|
5
|
0.125
|
0.078125
|
|
GO:BP
|
GO:0071677
|
positive regulation of mononuclear cell migration
|
4.33562876206046e-06
|
65
|
123
|
7
|
0.0569105691056911
|
0.107692307692308
|
|
GO:BP
|
GO:0046631
|
alpha-beta T cell activation
|
4.35323823906304e-06
|
155
|
52
|
7
|
0.134615384615385
|
0.0451612903225806
|
|
GO:BP
|
GO:0048518
|
positive regulation of biological process
|
4.47888260616763e-06
|
6363
|
55
|
38
|
0.690909090909091
|
0.00597202577400597
|
|
GO:BP
|
GO:0051250
|
negative regulation of lymphocyte activation
|
4.50491994569098e-06
|
156
|
52
|
7
|
0.134615384615385
|
0.0448717948717949
|
|
GO:BP
|
GO:0042267
|
natural killer cell mediated cytotoxicity
|
4.64827263622904e-06
|
62
|
42
|
5
|
0.119047619047619
|
0.0806451612903226
|
|
GO:BP
|
GO:1903706
|
regulation of hemopoiesis
|
5.4564333693207e-06
|
424
|
8
|
5
|
0.625
|
0.0117924528301887
|
|
GO:BP
|
GO:1990266
|
neutrophil migration
|
5.58416333956063e-06
|
124
|
135
|
9
|
0.0666666666666667
|
0.0725806451612903
|
|
GO:BP
|
GO:0051241
|
negative regulation of multicellular organismal process
|
5.6298616538862e-06
|
1160
|
135
|
26
|
0.192592592592593
|
0.0224137931034483
|
|
GO:BP
|
GO:1903131
|
mononuclear cell differentiation
|
5.76763433400737e-06
|
430
|
8
|
5
|
0.625
|
0.0116279069767442
|
|
GO:BP
|
GO:0002228
|
natural killer cell mediated immunity
|
5.76763433400737e-06
|
65
|
42
|
5
|
0.119047619047619
|
0.0769230769230769
|
|
GO:BP
|
GO:0048525
|
negative regulation of viral process
|
5.85914277648712e-06
|
94
|
131
|
8
|
0.0610687022900763
|
0.0851063829787234
|
|
GO:BP
|
GO:0071621
|
granulocyte chemotaxis
|
6.25410677725354e-06
|
126
|
135
|
9
|
0.0666666666666667
|
0.0714285714285714
|
|
GO:BP
|
GO:0046635
|
positive regulation of alpha-beta T cell activation
|
6.62947937031862e-06
|
67
|
42
|
5
|
0.119047619047619
|
0.0746268656716418
|
|
GO:BP
|
GO:0002584
|
negative regulation of antigen processing and presentation of peptide antigen
|
6.85793828388705e-06
|
4
|
90
|
3
|
0.0333333333333333
|
0.75
|
|
GO:BP
|
GO:0050866
|
negative regulation of cell activation
|
7.51316729213947e-06
|
211
|
135
|
11
|
0.0814814814814815
|
0.0521327014218009
|
|
GO:BP
|
GO:0002507
|
tolerance induction
|
8.13483797402989e-06
|
29
|
42
|
4
|
0.0952380952380952
|
0.137931034482759
|
|
GO:BP
|
GO:0045580
|
regulation of T cell differentiation
|
8.25547290809659e-06
|
152
|
3
|
3
|
1
|
0.0197368421052632
|
|
GO:BP
|
GO:0030217
|
T cell differentiation
|
8.62860955587552e-06
|
260
|
6
|
4
|
0.666666666666667
|
0.0153846153846154
|
|
GO:BP
|
GO:0050691
|
regulation of defense response to virus by host
|
8.78091746168267e-06
|
43
|
131
|
6
|
0.0458015267175573
|
0.13953488372093
|
|
GO:BP
|
GO:1902036
|
regulation of hematopoietic stem cell differentiation
|
8.78091746168267e-06
|
75
|
40
|
5
|
0.125
|
0.0666666666666667
|
|
GO:BP
|
GO:1902107
|
positive regulation of leukocyte differentiation
|
8.86760961100292e-06
|
157
|
3
|
3
|
1
|
0.0191082802547771
|
|
GO:BP
|
GO:1903708
|
positive regulation of hemopoiesis
|
8.86760961100292e-06
|
157
|
3
|
3
|
1
|
0.0191082802547771
|
|
GO:BP
|
GO:0140289
|
protein mono-ADP-ribosylation
|
8.86760961100292e-06
|
12
|
111
|
4
|
0.036036036036036
|
0.333333333333333
|
|
GO:BP
|
GO:0038061
|
NIK/NF-kappaB signaling
|
9.8539397020342e-06
|
194
|
69
|
8
|
0.115942028985507
|
0.0412371134020619
|
|
GO:BP
|
GO:0061418
|
regulation of transcription from RNA polymerase II promoter in response to hypoxia
|
1.04158057467962e-05
|
78
|
40
|
5
|
0.125
|
0.0641025641025641
|
|
GO:BP
|
GO:0000209
|
protein polyubiquitination
|
1.15555918059809e-05
|
342
|
127
|
13
|
0.102362204724409
|
0.0380116959064327
|
|
GO:BP
|
GO:0002687
|
positive regulation of leukocyte migration
|
1.17885419694377e-05
|
137
|
135
|
9
|
0.0666666666666667
|
0.0656934306569343
|
|
GO:BP
|
GO:0046718
|
viral entry into host cell
|
1.18756611463432e-05
|
141
|
67
|
7
|
0.104477611940299
|
0.049645390070922
|
|
GO:BP
|
GO:0045619
|
regulation of lymphocyte differentiation
|
1.30042778122281e-05
|
180
|
3
|
3
|
1
|
0.0166666666666667
|
|
GO:BP
|
GO:0033238
|
regulation of cellular amine metabolic process
|
1.30968946537167e-05
|
82
|
40
|
5
|
0.125
|
0.0609756097560976
|
|
GO:BP
|
GO:0098609
|
cell-cell adhesion
|
1.32606832554493e-05
|
894
|
30
|
10
|
0.333333333333333
|
0.0111856823266219
|
|
GO:BP
|
GO:0002695
|
negative regulation of leukocyte activation
|
1.38531066617705e-05
|
188
|
52
|
7
|
0.134615384615385
|
0.0372340425531915
|
|
GO:BP
|
GO:0030595
|
leukocyte chemotaxis
|
1.42012208653026e-05
|
227
|
135
|
11
|
0.0814814814814815
|
0.0484581497797357
|
|
GO:BP
|
GO:0002285
|
lymphocyte activation involved in immune response
|
1.4932955516901e-05
|
190
|
3
|
3
|
1
|
0.0157894736842105
|
|
GO:BP
|
GO:0031145
|
anaphase-promoting complex-dependent catabolic process
|
1.5286541513718e-05
|
85
|
40
|
5
|
0.125
|
0.0588235294117647
|
|
GO:BP
|
GO:0016045
|
detection of bacterium
|
1.55443389949197e-05
|
17
|
23
|
3
|
0.130434782608696
|
0.176470588235294
|
|
GO:BP
|
GO:0045589
|
regulation of regulatory T cell differentiation
|
1.59148053421637e-05
|
35
|
42
|
4
|
0.0952380952380952
|
0.114285714285714
|
|
GO:BP
|
GO:0010628
|
positive regulation of gene expression
|
1.59293931831087e-05
|
1160
|
30
|
11
|
0.366666666666667
|
0.00948275862068966
|
|
GO:BP
|
GO:0002521
|
leukocyte differentiation
|
1.67429901358629e-05
|
550
|
8
|
5
|
0.625
|
0.00909090909090909
|
|
GO:BP
|
GO:0050868
|
negative regulation of T cell activation
|
1.73295245081451e-05
|
123
|
52
|
6
|
0.115384615384615
|
0.0487804878048781
|
|
GO:BP
|
GO:0044106
|
cellular amine metabolic process
|
1.8140653924847e-05
|
163
|
40
|
6
|
0.15
|
0.0368098159509202
|
|
GO:BP
|
GO:1901532
|
regulation of hematopoietic progenitor cell differentiation
|
1.83256326600335e-05
|
89
|
40
|
5
|
0.125
|
0.0561797752808989
|
|
GO:BP
|
GO:0060218
|
hematopoietic stem cell differentiation
|
1.83256326600335e-05
|
89
|
40
|
5
|
0.125
|
0.0561797752808989
|
|
GO:BP
|
GO:0051240
|
positive regulation of multicellular organismal process
|
1.88773867958531e-05
|
1455
|
30
|
12
|
0.4
|
0.00824742268041237
|
|
GO:BP
|
GO:0044409
|
entry into host
|
1.96610476038679e-05
|
154
|
67
|
7
|
0.104477611940299
|
0.0454545454545455
|
|
GO:BP
|
GO:0098543
|
detection of other organism
|
2.10171995512431e-05
|
19
|
23
|
3
|
0.130434782608696
|
0.157894736842105
|
|
GO:BP
|
GO:0009308
|
amine metabolic process
|
2.10561593873676e-05
|
168
|
40
|
6
|
0.15
|
0.0357142857142857
|
|
GO:BP
|
GO:0045066
|
regulatory T cell differentiation
|
2.11801566912996e-05
|
38
|
42
|
4
|
0.0952380952380952
|
0.105263157894737
|
|
GO:BP
|
GO:0090713
|
immunological memory process
|
2.11801566912996e-05
|
17
|
97
|
4
|
0.0412371134020619
|
0.235294117647059
|
|
GO:BP
|
GO:0042180
|
cellular ketone metabolic process
|
2.29629031591355e-05
|
271
|
40
|
7
|
0.175
|
0.025830258302583
|
|
GO:BP
|
GO:0097530
|
granulocyte migration
|
2.36005385686154e-05
|
151
|
135
|
9
|
0.0666666666666667
|
0.0596026490066225
|
|
GO:BP
|
GO:0031146
|
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process
|
2.43892544612738e-05
|
95
|
40
|
5
|
0.125
|
0.0526315789473684
|
|
GO:BP
|
GO:0002583
|
regulation of antigen processing and presentation of peptide antigen
|
2.80951096291224e-05
|
6
|
90
|
3
|
0.0333333333333333
|
0.5
|
|
GO:BP
|
GO:0010565
|
regulation of cellular ketone metabolic process
|
2.82465189927981e-05
|
194
|
21
|
5
|
0.238095238095238
|
0.0257731958762887
|
|
GO:BP
|
GO:0006471
|
protein ADP-ribosylation
|
3.02634922515928e-05
|
36
|
111
|
5
|
0.045045045045045
|
0.138888888888889
|
|
GO:BP
|
GO:0045595
|
regulation of cell differentiation
|
3.11658932852796e-05
|
1679
|
42
|
15
|
0.357142857142857
|
0.00893388921977368
|
|
GO:BP
|
GO:0030098
|
lymphocyte differentiation
|
3.11658932852796e-05
|
376
|
6
|
4
|
0.666666666666667
|
0.0106382978723404
|
|
GO:BP
|
GO:0010972
|
negative regulation of G2/M transition of mitotic cell cycle
|
3.37847110166763e-05
|
102
|
40
|
5
|
0.125
|
0.0490196078431373
|
|
GO:BP
|
GO:2001185
|
regulation of CD8-positive, alpha-beta T cell activation
|
3.4564015456367e-05
|
18
|
29
|
3
|
0.103448275862069
|
0.166666666666667
|
|
GO:BP
|
GO:0032102
|
negative regulation of response to external stimulus
|
3.47934093480413e-05
|
406
|
69
|
10
|
0.144927536231884
|
0.0246305418719212
|
|
GO:BP
|
GO:1903038
|
negative regulation of leukocyte cell-cell adhesion
|
3.6574509995334e-05
|
142
|
52
|
6
|
0.115384615384615
|
0.0422535211267606
|
|
GO:BP
|
GO:0002577
|
regulation of antigen processing and presentation
|
3.68932916776146e-05
|
21
|
90
|
4
|
0.0444444444444444
|
0.19047619047619
|
|
GO:BP
|
GO:0002690
|
positive regulation of leukocyte chemotaxis
|
3.76443551564369e-05
|
93
|
123
|
7
|
0.0569105691056911
|
0.0752688172043011
|
|
GO:BP
|
GO:0070498
|
interleukin-1-mediated signaling pathway
|
3.81456793369194e-05
|
105
|
40
|
5
|
0.125
|
0.0476190476190476
|
|
GO:BP
|
GO:0010468
|
regulation of gene expression
|
3.91735784959518e-05
|
5196
|
24
|
18
|
0.75
|
0.00346420323325635
|
|
GO:BP
|
GO:0098581
|
detection of external biotic stimulus
|
4.04741635565073e-05
|
24
|
23
|
3
|
0.130434782608696
|
0.125
|
|
GO:BP
|
GO:0002218
|
activation of innate immune response
|
4.349657447554e-05
|
144
|
53
|
6
|
0.113207547169811
|
0.0416666666666667
|
|
GO:BP
|
GO:0070098
|
chemokine-mediated signaling pathway
|
4.38963479133252e-05
|
87
|
135
|
7
|
0.0518518518518519
|
0.0804597701149425
|
|
GO:BP
|
GO:1902105
|
regulation of leukocyte differentiation
|
4.47932924724958e-05
|
288
|
3
|
3
|
1
|
0.0104166666666667
|
|
GO:BP
|
GO:0052126
|
movement in host environment
|
4.49540811949961e-05
|
177
|
67
|
7
|
0.104477611940299
|
0.0395480225988701
|
|
GO:BP
|
GO:0060071
|
Wnt signaling pathway, planar cell polarity pathway
|
4.67962088177041e-05
|
110
|
40
|
5
|
0.125
|
0.0454545454545455
|
|
GO:BP
|
GO:0048522
|
positive regulation of cellular process
|
4.76774480541676e-05
|
5757
|
55
|
34
|
0.618181818181818
|
0.00590585374326906
|
|
GO:BP
|
GO:0043618
|
regulation of transcription from RNA polymerase II promoter in response to stress
|
4.84202856409392e-05
|
111
|
40
|
5
|
0.125
|
0.045045045045045
|
|
GO:BP
|
GO:0002223
|
stimulatory C-type lectin receptor signaling pathway
|
5.04280059000813e-05
|
112
|
40
|
5
|
0.125
|
0.0446428571428571
|
|
GO:BP
|
GO:1902750
|
negative regulation of cell cycle G2/M phase transition
|
5.24978817396877e-05
|
113
|
40
|
5
|
0.125
|
0.0442477876106195
|
|
GO:BP
|
GO:0090175
|
regulation of establishment of planar polarity
|
5.46311686058453e-05
|
114
|
40
|
5
|
0.125
|
0.043859649122807
|
|
GO:BP
|
GO:2000736
|
regulation of stem cell differentiation
|
5.66371423712639e-05
|
115
|
40
|
5
|
0.125
|
0.0434782608695652
|
|
GO:BP
|
GO:0002220
|
innate immune response activating cell surface receptor signaling pathway
|
5.66371423712639e-05
|
115
|
40
|
5
|
0.125
|
0.0434782608695652
|
|
GO:BP
|
GO:0032649
|
regulation of interferon-gamma production
|
5.81009693930305e-05
|
110
|
42
|
5
|
0.119047619047619
|
0.0454545454545455
|
|
GO:BP
|
GO:0002758
|
innate immune response-activating signal transduction
|
5.86964531568556e-05
|
116
|
40
|
5
|
0.125
|
0.0431034482758621
|
|
GO:BP
|
GO:0016064
|
immunoglobulin mediated immune response
|
5.92899915733769e-05
|
223
|
132
|
10
|
0.0757575757575758
|
0.0448430493273543
|
|
GO:BP
|
GO:2000403
|
positive regulation of lymphocyte migration
|
5.98910444317546e-05
|
38
|
123
|
5
|
0.040650406504065
|
0.131578947368421
|
|
GO:BP
|
GO:0043620
|
regulation of DNA-templated transcription in response to stress
|
6.06079418182168e-05
|
117
|
40
|
5
|
0.125
|
0.0427350427350427
|
|
GO:BP
|
GO:0048519
|
negative regulation of biological process
|
6.5921676084277e-05
|
5757
|
69
|
40
|
0.579710144927536
|
0.00694806322737537
|
|
GO:BP
|
GO:0019724
|
B cell mediated immunity
|
6.5921676084277e-05
|
226
|
132
|
10
|
0.0757575757575758
|
0.0442477876106195
|
|
GO:BP
|
GO:0097529
|
myeloid leukocyte migration
|
6.5921676084277e-05
|
221
|
135
|
10
|
0.0740740740740741
|
0.0452488687782805
|
|
GO:BP
|
GO:0050798
|
activated T cell proliferation
|
6.62841950300816e-05
|
49
|
97
|
5
|
0.0515463917525773
|
0.102040816326531
|
|
GO:BP
|
GO:0032020
|
ISG15-protein conjugation
|
6.97633034524726e-05
|
6
|
127
|
3
|
0.0236220472440945
|
0.5
|
|
GO:BP
|
GO:0006508
|
proteolysis
|
7.45873179895327e-05
|
1836
|
73
|
21
|
0.287671232876712
|
0.011437908496732
|
|
GO:BP
|
GO:0048247
|
lymphocyte chemotaxis
|
7.68091862691338e-05
|
63
|
135
|
6
|
0.0444444444444444
|
0.0952380952380952
|
|
GO:BP
|
GO:1990868
|
response to chemokine
|
8.34083215009384e-05
|
97
|
135
|
7
|
0.0518518518518519
|
0.0721649484536082
|
|
GO:BP
|
GO:1990869
|
cellular response to chemokine
|
8.34083215009384e-05
|
97
|
135
|
7
|
0.0518518518518519
|
0.0721649484536082
|
|
GO:BP
|
GO:0006520
|
cellular amino acid metabolic process
|
8.90880267219259e-05
|
341
|
40
|
7
|
0.175
|
0.0205278592375367
|
|
GO:BP
|
GO:0048523
|
negative regulation of cellular process
|
9.08906068635479e-05
|
5139
|
69
|
37
|
0.536231884057971
|
0.00719984432769021
|
|
GO:BP
|
GO:0001736
|
establishment of planar polarity
|
9.65833470921914e-05
|
130
|
40
|
5
|
0.125
|
0.0384615384615385
|
|
GO:BP
|
GO:0007164
|
establishment of tissue polarity
|
9.65833470921914e-05
|
130
|
40
|
5
|
0.125
|
0.0384615384615385
|
|
GO:BP
|
GO:0002724
|
regulation of T cell cytokine production
|
9.66789167142136e-05
|
33
|
23
|
3
|
0.130434782608696
|
0.0909090909090909
|
|
GO:BP
|
GO:0002578
|
negative regulation of antigen processing and presentation
|
9.8743199784415e-05
|
9
|
90
|
3
|
0.0333333333333333
|
0.333333333333333
|
|
GO:BP
|
GO:0036037
|
CD8-positive, alpha-beta T cell activation
|
0.000105830599204929
|
27
|
29
|
3
|
0.103448275862069
|
0.111111111111111
|
|
GO:BP
|
GO:0032615
|
interleukin-12 production
|
0.000110788873077138
|
60
|
42
|
4
|
0.0952380952380952
|
0.0666666666666667
|
|
GO:BP
|
GO:0032655
|
regulation of interleukin-12 production
|
0.000110788873077138
|
60
|
42
|
4
|
0.0952380952380952
|
0.0666666666666667
|
|
GO:BP
|
GO:0002274
|
myeloid leukocyte activation
|
0.00011557364986198
|
667
|
50
|
10
|
0.2
|
0.0149925037481259
|
|
GO:BP
|
GO:0071675
|
regulation of mononuclear cell migration
|
0.000118578923210878
|
113
|
123
|
7
|
0.0569105691056911
|
0.0619469026548673
|
|
GO:BP
|
GO:0070212
|
protein poly-ADP-ribosylation
|
0.00012152300886302
|
8
|
111
|
3
|
0.027027027027027
|
0.375
|
|
GO:BP
|
GO:0009595
|
detection of biotic stimulus
|
0.000121972541933967
|
36
|
23
|
3
|
0.130434782608696
|
0.0833333333333333
|
|
GO:BP
|
GO:0002369
|
T cell cytokine production
|
0.000121972541933967
|
36
|
23
|
3
|
0.130434782608696
|
0.0833333333333333
|
|
GO:BP
|
GO:0050688
|
regulation of defense response to virus
|
0.000121972541933967
|
71
|
131
|
6
|
0.0458015267175573
|
0.0845070422535211
|
|
GO:BP
|
GO:0008284
|
positive regulation of cell population proliferation
|
0.000126620611152936
|
982
|
43
|
11
|
0.255813953488372
|
0.0112016293279022
|
|
GO:BP
|
GO:0042832
|
defense response to protozoan
|
0.000128311445912043
|
25
|
107
|
4
|
0.0373831775700935
|
0.16
|
|
GO:BP
|
GO:0071674
|
mononuclear cell migration
|
0.00012857463136009
|
191
|
135
|
9
|
0.0666666666666667
|
0.0471204188481675
|
|
GO:BP
|
GO:0046641
|
positive regulation of alpha-beta T cell proliferation
|
0.000143950136703481
|
21
|
42
|
3
|
0.0714285714285714
|
0.142857142857143
|
|
GO:BP
|
GO:0001562
|
response to protozoan
|
0.00014915601789802
|
26
|
107
|
4
|
0.0373831775700935
|
0.153846153846154
|
|
GO:BP
|
GO:0032729
|
positive regulation of interferon-gamma production
|
0.000153976129072188
|
66
|
42
|
4
|
0.0952380952380952
|
0.0606060606060606
|
|
GO:BP
|
GO:0010499
|
proteasomal ubiquitin-independent protein catabolic process
|
0.000161808639098069
|
23
|
40
|
3
|
0.075
|
0.130434782608696
|
|
GO:BP
|
GO:0051701
|
biological process involved in interaction with host
|
0.000161808639098069
|
221
|
67
|
7
|
0.104477611940299
|
0.0316742081447964
|
|
GO:BP
|
GO:0002688
|
regulation of leukocyte chemotaxis
|
0.000166393838052238
|
120
|
123
|
7
|
0.0569105691056911
|
0.0583333333333333
|
|
GO:BP
|
GO:0022408
|
negative regulation of cell-cell adhesion
|
0.00017664432001337
|
194
|
52
|
6
|
0.115384615384615
|
0.0309278350515464
|
|
GO:BP
|
GO:0010950
|
positive regulation of endopeptidase activity
|
0.000178373068776536
|
187
|
54
|
6
|
0.111111111111111
|
0.0320855614973262
|
|
GO:BP
|
GO:0035567
|
non-canonical Wnt signaling pathway
|
0.000181751537759092
|
151
|
40
|
5
|
0.125
|
0.033112582781457
|
|
GO:BP
|
GO:0090263
|
positive regulation of canonical Wnt signaling pathway
|
0.000181751537759092
|
151
|
40
|
5
|
0.125
|
0.033112582781457
|
|
GO:BP
|
GO:0001738
|
morphogenesis of a polarized epithelium
|
0.000187091772000612
|
152
|
40
|
5
|
0.125
|
0.0328947368421053
|
|
GO:BP
|
GO:0009967
|
positive regulation of signal transduction
|
0.000191533144844148
|
1598
|
69
|
18
|
0.260869565217391
|
0.0112640801001252
|
|
GO:BP
|
GO:0060326
|
cell chemotaxis
|
0.000193311715843802
|
310
|
135
|
11
|
0.0814814814814815
|
0.0354838709677419
|
|
GO:BP
|
GO:0030162
|
regulation of proteolysis
|
0.00020501305262086
|
752
|
58
|
11
|
0.189655172413793
|
0.0146276595744681
|
|
GO:BP
|
GO:0010467
|
gene expression
|
0.000205716441098314
|
6179
|
57
|
35
|
0.614035087719298
|
0.00566434698171225
|
|
GO:BP
|
GO:0010647
|
positive regulation of cell communication
|
0.000215170099083443
|
1777
|
69
|
19
|
0.27536231884058
|
0.0106921778277997
|
|
GO:BP
|
GO:0007162
|
negative regulation of cell adhesion
|
0.000219155150987266
|
304
|
52
|
7
|
0.134615384615385
|
0.0230263157894737
|
|
GO:BP
|
GO:0001818
|
negative regulation of cytokine production
|
0.000219765236797544
|
381
|
42
|
7
|
0.166666666666667
|
0.0183727034120735
|
|
GO:BP
|
GO:0023056
|
positive regulation of signaling
|
0.000221350350558594
|
1782
|
69
|
19
|
0.27536231884058
|
0.010662177328844
|
|
GO:BP
|
GO:0006954
|
inflammatory response
|
0.000222483148324952
|
794
|
135
|
18
|
0.133333333333333
|
0.0226700251889169
|
|
GO:BP
|
GO:0042130
|
negative regulation of T cell proliferation
|
0.000236988776977601
|
70
|
91
|
5
|
0.0549450549450549
|
0.0714285714285714
|
|
GO:BP
|
GO:1901623
|
regulation of lymphocyte chemotaxis
|
0.000237819664173812
|
26
|
123
|
4
|
0.032520325203252
|
0.153846153846154
|
|
GO:BP
|
GO:0072676
|
lymphocyte migration
|
0.000241083802144658
|
117
|
135
|
7
|
0.0518518518518519
|
0.0598290598290598
|
|
GO:BP
|
GO:0010629
|
negative regulation of gene expression
|
0.000243827157465343
|
1460
|
44
|
13
|
0.295454545454545
|
0.0089041095890411
|
|
GO:BP
|
GO:0050830
|
defense response to Gram-positive bacterium
|
0.000246078720172426
|
101
|
106
|
6
|
0.0566037735849057
|
0.0594059405940594
|
|
GO:BP
|
GO:0002685
|
regulation of leukocyte migration
|
0.0002555373946987
|
211
|
135
|
9
|
0.0666666666666667
|
0.042654028436019
|
|
GO:BP
|
GO:0009435
|
NAD biosynthetic process
|
0.000263074313880918
|
29
|
113
|
4
|
0.0353982300884956
|
0.137931034482759
|
|
GO:BP
|
GO:0035458
|
cellular response to interferon-beta
|
0.000263074313880918
|
21
|
53
|
3
|
0.0566037735849057
|
0.142857142857143
|
|
GO:BP
|
GO:0038095
|
Fc-epsilon receptor signaling pathway
|
0.00027441453874498
|
167
|
40
|
5
|
0.125
|
0.029940119760479
|
|
GO:BP
|
GO:0043299
|
leukocyte degranulation
|
0.000275381940778412
|
535
|
43
|
8
|
0.186046511627907
|
0.0149532710280374
|
|
GO:BP
|
GO:0010952
|
positive regulation of peptidase activity
|
0.000275381940778412
|
205
|
54
|
6
|
0.111111111111111
|
0.0292682926829268
|
|
GO:BP
|
GO:0045071
|
negative regulation of viral genome replication
|
0.000275630159852389
|
54
|
123
|
5
|
0.040650406504065
|
0.0925925925925926
|
|
GO:BP
|
GO:0051603
|
proteolysis involved in cellular protein catabolic process
|
0.000308542689763666
|
750
|
41
|
9
|
0.219512195121951
|
0.012
|
|
GO:BP
|
GO:0010818
|
T cell chemotaxis
|
0.00031021483092498
|
28
|
123
|
4
|
0.032520325203252
|
0.142857142857143
|
|
GO:BP
|
GO:0002244
|
hematopoietic progenitor cell differentiation
|
0.00031021483092498
|
172
|
40
|
5
|
0.125
|
0.0290697674418605
|
|
GO:BP
|
GO:0002444
|
myeloid leukocyte mediated immunity
|
0.00033269100435466
|
551
|
43
|
8
|
0.186046511627907
|
0.014519056261343
|
|
GO:BP
|
GO:0007155
|
cell adhesion
|
0.00033833261650464
|
1492
|
33
|
11
|
0.333333333333333
|
0.00737265415549598
|
|
GO:BP
|
GO:0051259
|
protein complex oligomerization
|
0.000345871511943052
|
234
|
6
|
3
|
0.5
|
0.0128205128205128
|
|
GO:BP
|
GO:0022610
|
biological adhesion
|
0.000347871151041293
|
1499
|
33
|
11
|
0.333333333333333
|
0.00733822548365577
|
|
GO:BP
|
GO:0050793
|
regulation of developmental process
|
0.000347871151041293
|
2605
|
55
|
20
|
0.363636363636364
|
0.00767754318618042
|
|
GO:BP
|
GO:0019359
|
nicotinamide nucleotide biosynthetic process
|
0.000369662442624517
|
32
|
113
|
4
|
0.0353982300884956
|
0.125
|
|
GO:BP
|
GO:0019363
|
pyridine nucleotide biosynthetic process
|
0.000369662442624517
|
32
|
113
|
4
|
0.0353982300884956
|
0.125
|
|
GO:BP
|
GO:0050921
|
positive regulation of chemotaxis
|
0.000406989392344695
|
141
|
123
|
7
|
0.0569105691056911
|
0.049645390070922
|
|
GO:BP
|
GO:0032673
|
regulation of interleukin-4 production
|
0.000406989392344695
|
31
|
42
|
3
|
0.0714285714285714
|
0.0967741935483871
|
|
GO:BP
|
GO:2000027
|
regulation of animal organ morphogenesis
|
0.000409580984975881
|
184
|
40
|
5
|
0.125
|
0.0271739130434783
|
|
GO:BP
|
GO:0030177
|
positive regulation of Wnt signaling pathway
|
0.000409580984975881
|
184
|
40
|
5
|
0.125
|
0.0271739130434783
|
|
GO:BP
|
GO:0019730
|
antimicrobial humoral response
|
0.000432474107854476
|
145
|
121
|
7
|
0.0578512396694215
|
0.0482758620689655
|
|
GO:BP
|
GO:0090090
|
negative regulation of canonical Wnt signaling pathway
|
0.000437955386462327
|
187
|
40
|
5
|
0.125
|
0.0267379679144385
|
|
GO:BP
|
GO:0032633
|
interleukin-4 production
|
0.000439492949725405
|
32
|
42
|
3
|
0.0714285714285714
|
0.09375
|
|
GO:BP
|
GO:2000406
|
positive regulation of T cell migration
|
0.000440505764106314
|
31
|
123
|
4
|
0.032520325203252
|
0.129032258064516
|
|
GO:BP
|
GO:0045637
|
regulation of myeloid cell differentiation
|
0.000464780608272861
|
263
|
6
|
3
|
0.5
|
0.0114068441064639
|
|
GO:BP
|
GO:0043488
|
regulation of mRNA stability
|
0.000477126543271693
|
191
|
40
|
5
|
0.125
|
0.0261780104712042
|
|
GO:BP
|
GO:0072525
|
pyridine-containing compound biosynthetic process
|
0.000508552217979133
|
35
|
113
|
4
|
0.0353982300884956
|
0.114285714285714
|
|
GO:BP
|
GO:0046496
|
nicotinamide nucleotide metabolic process
|
0.000508552217979133
|
35
|
113
|
4
|
0.0353982300884956
|
0.114285714285714
|
|
GO:BP
|
GO:0019362
|
pyridine nucleotide metabolic process
|
0.000508552217979133
|
35
|
113
|
4
|
0.0353982300884956
|
0.114285714285714
|
|
GO:BP
|
GO:0046007
|
negative regulation of activated T cell proliferation
|
0.000512058423679196
|
16
|
91
|
3
|
0.032967032967033
|
0.1875
|
|
GO:BP
|
GO:0044257
|
cellular protein catabolic process
|
0.000528571382597081
|
813
|
41
|
9
|
0.219512195121951
|
0.011070110701107
|
|
GO:BP
|
GO:2000401
|
regulation of lymphocyte migration
|
0.000530772694191595
|
63
|
123
|
5
|
0.040650406504065
|
0.0793650793650794
|
|
GO:BP
|
GO:0050672
|
negative regulation of lymphocyte proliferation
|
0.000530988085708498
|
85
|
91
|
5
|
0.0549450549450549
|
0.0588235294117647
|
|
GO:BP
|
GO:0032945
|
negative regulation of mononuclear cell proliferation
|
0.000560197152947413
|
86
|
91
|
5
|
0.0549450549450549
|
0.0581395348837209
|
|
GO:BP
|
GO:0006511
|
ubiquitin-dependent protein catabolic process
|
0.000573399336623465
|
655
|
21
|
6
|
0.285714285714286
|
0.00916030534351145
|
|
GO:BP
|
GO:0043487
|
regulation of RNA stability
|
0.000582278928552782
|
201
|
21
|
4
|
0.19047619047619
|
0.0199004975124378
|
|
GO:BP
|
GO:0009968
|
negative regulation of signal transduction
|
0.000587184127644065
|
1364
|
58
|
14
|
0.241379310344828
|
0.0102639296187683
|
|
GO:BP
|
GO:0019941
|
modification-dependent protein catabolic process
|
0.000602450561375841
|
662
|
21
|
6
|
0.285714285714286
|
0.00906344410876133
|
|
GO:BP
|
GO:0016567
|
protein ubiquitination
|
0.00063880664840897
|
892
|
23
|
7
|
0.304347826086957
|
0.007847533632287
|
|
GO:BP
|
GO:0010389
|
regulation of G2/M transition of mitotic cell cycle
|
0.000646441326584994
|
207
|
21
|
4
|
0.19047619047619
|
0.0193236714975845
|
|
GO:BP
|
GO:0043632
|
modification-dependent macromolecule catabolic process
|
0.00065536363806301
|
673
|
21
|
6
|
0.285714285714286
|
0.00891530460624071
|
|
GO:BP
|
GO:0072678
|
T cell migration
|
0.000693576264478385
|
67
|
123
|
5
|
0.040650406504065
|
0.0746268656716418
|
|
GO:BP
|
GO:0071456
|
cellular response to hypoxia
|
0.000704034145009699
|
212
|
21
|
4
|
0.19047619047619
|
0.0188679245283019
|
|
GO:BP
|
GO:0061013
|
regulation of mRNA catabolic process
|
0.000715293038380329
|
213
|
21
|
4
|
0.19047619047619
|
0.0187793427230047
|
|
GO:BP
|
GO:0046006
|
regulation of activated T cell proliferation
|
0.000728484844765351
|
45
|
97
|
4
|
0.0412371134020619
|
0.0888888888888889
|
|
GO:BP
|
GO:0032735
|
positive regulation of interleukin-12 production
|
0.00073241326807009
|
41
|
107
|
4
|
0.0373831775700935
|
0.0975609756097561
|
|
GO:BP
|
GO:0030890
|
positive regulation of B cell proliferation
|
0.00073241326807009
|
41
|
107
|
4
|
0.0373831775700935
|
0.0975609756097561
|
|
GO:BP
|
GO:0070664
|
negative regulation of leukocyte proliferation
|
0.000740509408658349
|
92
|
91
|
5
|
0.0549450549450549
|
0.0543478260869565
|
|
GO:BP
|
GO:0030178
|
negative regulation of Wnt signaling pathway
|
0.000769828407925241
|
218
|
21
|
4
|
0.19047619047619
|
0.018348623853211
|
|
GO:BP
|
GO:0070234
|
positive regulation of T cell apoptotic process
|
0.000777756014897941
|
14
|
123
|
3
|
0.024390243902439
|
0.214285714285714
|
|
GO:BP
|
GO:0036294
|
cellular response to decreased oxygen levels
|
0.000820170167271053
|
222
|
21
|
4
|
0.19047619047619
|
0.018018018018018
|
|
GO:BP
|
GO:1903901
|
negative regulation of viral life cycle
|
0.000829913514955734
|
26
|
66
|
3
|
0.0454545454545455
|
0.115384615384615
|
|
GO:BP
|
GO:1902749
|
regulation of cell cycle G2/M phase transition
|
0.000830728214396488
|
223
|
21
|
4
|
0.19047619047619
|
0.0179372197309417
|
|
GO:BP
|
GO:0045597
|
positive regulation of cell differentiation
|
0.000831731206045017
|
879
|
3
|
3
|
1
|
0.00341296928327645
|
|
GO:BP
|
GO:0043161
|
proteasome-mediated ubiquitin-dependent protein catabolic process
|
0.000832723201190672
|
434
|
21
|
5
|
0.238095238095238
|
0.0115207373271889
|
|
GO:BP
|
GO:0002275
|
myeloid cell activation involved in immune response
|
0.00083608173946721
|
547
|
50
|
8
|
0.16
|
0.0146252285191956
|
|
GO:BP
|
GO:2000116
|
regulation of cysteine-type endopeptidase activity
|
0.000836507625418334
|
240
|
58
|
6
|
0.103448275862069
|
0.025
|
|
GO:BP
|
GO:0072524
|
pyridine-containing compound metabolic process
|
0.000878533141685149
|
41
|
113
|
4
|
0.0353982300884956
|
0.0975609756097561
|
|
GO:BP
|
GO:0060334
|
regulation of interferon-gamma-mediated signaling pathway
|
0.000888962074645878
|
26
|
68
|
3
|
0.0441176470588235
|
0.115384615384615
|
|
GO:BP
|
GO:0060330
|
regulation of response to interferon-gamma
|
0.000888962074645878
|
26
|
68
|
3
|
0.0441176470588235
|
0.115384615384615
|
|
GO:BP
|
GO:0022603
|
regulation of anatomical structure morphogenesis
|
0.000915635626318363
|
1042
|
43
|
10
|
0.232558139534884
|
0.00959692898272553
|
|
GO:BP
|
GO:0062012
|
regulation of small molecule metabolic process
|
0.0009394825784959
|
447
|
21
|
5
|
0.238095238095238
|
0.0111856823266219
|
|
GO:BP
|
GO:0043687
|
post-translational protein modification
|
0.000943068655201285
|
362
|
40
|
6
|
0.15
|
0.0165745856353591
|
|
GO:BP
|
GO:0035455
|
response to interferon-alpha
|
0.000946018131076426
|
23
|
79
|
3
|
0.0379746835443038
|
0.130434782608696
|
|
GO:BP
|
GO:0055065
|
metal ion homeostasis
|
0.000968044321503979
|
667
|
135
|
15
|
0.111111111111111
|
0.0224887556221889
|
|
GO:BP
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.000969140986236477
|
6603
|
24
|
18
|
0.75
|
0.00272603362108133
|
|
GO:BP
|
GO:0006875
|
cellular metal ion homeostasis
|
0.000983838454769414
|
593
|
135
|
14
|
0.103703703703704
|
0.0236087689713322
|
|
GO:BP
|
GO:0002548
|
monocyte chemotaxis
|
0.00100259940275145
|
67
|
135
|
5
|
0.037037037037037
|
0.0746268656716418
|
|
GO:BP
|
GO:0032446
|
protein modification by small protein conjugation
|
0.00101814303266059
|
973
|
23
|
7
|
0.304347826086957
|
0.00719424460431655
|
|
GO:BP
|
GO:0038093
|
Fc receptor signaling pathway
|
0.00102112247693493
|
238
|
21
|
4
|
0.19047619047619
|
0.0168067226890756
|
|
GO:BP
|
GO:2000026
|
regulation of multicellular organismal development
|
0.00102920277192649
|
1447
|
8
|
5
|
0.625
|
0.00345542501727713
|
|
GO:BP
|
GO:0071453
|
cellular response to oxygen levels
|
0.00104994960064165
|
240
|
21
|
4
|
0.19047619047619
|
0.0166666666666667
|
|
GO:BP
|
GO:0008285
|
negative regulation of cell population proliferation
|
0.0010734122061088
|
790
|
91
|
13
|
0.142857142857143
|
0.0164556962025316
|
|
GO:BP
|
GO:0045055
|
regulated exocytosis
|
0.00110548631451928
|
785
|
47
|
9
|
0.191489361702128
|
0.0114649681528662
|
|
GO:BP
|
GO:0010648
|
negative regulation of cell communication
|
0.00114933642190028
|
1468
|
58
|
14
|
0.241379310344828
|
0.00953678474114441
|
|
GO:BP
|
GO:0023057
|
negative regulation of signaling
|
0.00117181884683935
|
1471
|
58
|
14
|
0.241379310344828
|
0.00951733514615908
|
|
GO:BP
|
GO:0046903
|
secretion
|
0.00119298827149154
|
1501
|
8
|
5
|
0.625
|
0.0033311125916056
|
|
GO:BP
|
GO:0097028
|
dendritic cell differentiation
|
0.00126399469783538
|
48
|
42
|
3
|
0.0714285714285714
|
0.0625
|
|
GO:BP
|
GO:0010819
|
regulation of T cell chemotaxis
|
0.0013029719436909
|
17
|
123
|
3
|
0.024390243902439
|
0.176470588235294
|
|
GO:BP
|
GO:0070230
|
positive regulation of lymphocyte apoptotic process
|
0.0013029719436909
|
17
|
123
|
3
|
0.024390243902439
|
0.176470588235294
|
|
GO:BP
|
GO:0048863
|
stem cell differentiation
|
0.00133375581649448
|
258
|
21
|
4
|
0.19047619047619
|
0.0155038759689922
|
|
GO:BP
|
GO:0002009
|
morphogenesis of an epithelium
|
0.00133605110989485
|
560
|
40
|
7
|
0.175
|
0.0125
|
|
GO:BP
|
GO:1901565
|
organonitrogen compound catabolic process
|
0.00133994381302932
|
1358
|
56
|
13
|
0.232142857142857
|
0.00957290132547865
|
|
GO:BP
|
GO:0010498
|
proteasomal protein catabolic process
|
0.0013572587237326
|
491
|
21
|
5
|
0.238095238095238
|
0.010183299389002
|
|
GO:BP
|
GO:0061844
|
antimicrobial humoral immune response mediated by antimicrobial peptide
|
0.0013911734015358
|
81
|
121
|
5
|
0.0413223140495868
|
0.0617283950617284
|
|
GO:BP
|
GO:1901991
|
negative regulation of mitotic cell cycle phase transition
|
0.00139718479621641
|
262
|
21
|
4
|
0.19047619047619
|
0.0152671755725191
|
|
GO:BP
|
GO:0000086
|
G2/M transition of mitotic cell cycle
|
0.00145636761145655
|
265
|
21
|
4
|
0.19047619047619
|
0.0150943396226415
|
|
GO:BP
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.00146390911891976
|
3241
|
69
|
25
|
0.36231884057971
|
0.00771366862079605
|
|
GO:BP
|
GO:2000404
|
regulation of T cell migration
|
0.00146677514528348
|
44
|
123
|
4
|
0.032520325203252
|
0.0909090909090909
|
|
GO:BP
|
GO:0000165
|
MAPK cascade
|
0.00156322560049187
|
935
|
42
|
9
|
0.214285714285714
|
0.00962566844919786
|
|
GO:BP
|
GO:0030099
|
myeloid cell differentiation
|
0.00161964175155539
|
428
|
6
|
3
|
0.5
|
0.00700934579439252
|
|
GO:BP
|
GO:0051281
|
positive regulation of release of sequestered calcium ion into cytosol
|
0.00172126179615855
|
42
|
135
|
4
|
0.0296296296296296
|
0.0952380952380952
|
|
GO:BP
|
GO:0030163
|
protein catabolic process
|
0.00172841120611912
|
974
|
41
|
9
|
0.219512195121951
|
0.00924024640657084
|
|
GO:BP
|
GO:0045069
|
regulation of viral genome replication
|
0.0017302241240002
|
84
|
123
|
5
|
0.040650406504065
|
0.0595238095238095
|
|
GO:BP
|
GO:0019079
|
viral genome replication
|
0.00175118358121258
|
131
|
123
|
6
|
0.0487804878048781
|
0.0458015267175573
|
|
GO:BP
|
GO:0002544
|
chronic inflammatory response
|
0.00176119488991517
|
19
|
123
|
3
|
0.024390243902439
|
0.157894736842105
|
|
GO:BP
|
GO:1901988
|
negative regulation of cell cycle phase transition
|
0.0017671133394902
|
281
|
21
|
4
|
0.19047619047619
|
0.0142348754448399
|
|
GO:BP
|
GO:0070228
|
regulation of lymphocyte apoptotic process
|
0.00179022924774296
|
55
|
42
|
3
|
0.0714285714285714
|
0.0545454545454545
|
|
GO:BP
|
GO:0044839
|
cell cycle G2/M phase transition
|
0.00180829492297573
|
283
|
21
|
4
|
0.19047619047619
|
0.0141342756183746
|
|
GO:BP
|
GO:0016579
|
protein deubiquitination
|
0.00200910938577623
|
292
|
21
|
4
|
0.19047619047619
|
0.0136986301369863
|
|
GO:BP
|
GO:0090026
|
positive regulation of monocyte chemotaxis
|
0.00201342512679225
|
20
|
123
|
3
|
0.024390243902439
|
0.15
|
|
GO:BP
|
GO:0050794
|
regulation of cellular process
|
0.0021169847857708
|
11473
|
58
|
49
|
0.844827586206897
|
0.00427089688834655
|
|
GO:BP
|
GO:0050801
|
ion homeostasis
|
0.00213361161447136
|
808
|
135
|
16
|
0.118518518518519
|
0.0198019801980198
|
|
GO:BP
|
GO:0060828
|
regulation of canonical Wnt signaling pathway
|
0.00214527901350032
|
298
|
21
|
4
|
0.19047619047619
|
0.0134228187919463
|
|
GO:BP
|
GO:0051928
|
positive regulation of calcium ion transport
|
0.00216312427085693
|
125
|
135
|
6
|
0.0444444444444444
|
0.048
|
|
GO:BP
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
0.00220377559034018
|
3483
|
32
|
15
|
0.46875
|
0.00430663221360896
|
|
GO:BP
|
GO:0002377
|
immunoglobulin production
|
0.00221132157110178
|
99
|
111
|
5
|
0.045045045045045
|
0.0505050505050505
|
|
GO:BP
|
GO:0048245
|
eosinophil chemotaxis
|
0.00222190920972839
|
19
|
135
|
3
|
0.0222222222222222
|
0.157894736842105
|
|
GO:BP
|
GO:0032653
|
regulation of interleukin-10 production
|
0.00223207185289809
|
60
|
42
|
3
|
0.0714285714285714
|
0.05
|
|
GO:BP
|
GO:0002224
|
toll-like receptor signaling pathway
|
0.00227969375856392
|
161
|
69
|
5
|
0.072463768115942
|
0.031055900621118
|
|
GO:BP
|
GO:0140131
|
positive regulation of lymphocyte chemotaxis
|
0.00228385506105641
|
21
|
123
|
3
|
0.024390243902439
|
0.142857142857143
|
|
GO:BP
|
GO:0006935
|
chemotaxis
|
0.00228779985278354
|
653
|
135
|
14
|
0.103703703703704
|
0.0214395099540582
|
|
GO:BP
|
GO:0002237
|
response to molecule of bacterial origin
|
0.00231294140801272
|
361
|
135
|
10
|
0.0740740740740741
|
0.0277008310249307
|
|
GO:BP
|
GO:0042330
|
taxis
|
0.00236812576297883
|
656
|
135
|
14
|
0.103703703703704
|
0.0213414634146341
|
|
GO:BP
|
GO:0051094
|
positive regulation of developmental process
|
0.00240478281765727
|
1341
|
6
|
4
|
0.666666666666667
|
0.00298284862043251
|
|
GO:BP
|
GO:0032613
|
interleukin-10 production
|
0.00240706084599583
|
62
|
42
|
3
|
0.0714285714285714
|
0.0483870967741935
|
|
GO:BP
|
GO:0070646
|
protein modification by small protein removal
|
0.00240775954350009
|
310
|
21
|
4
|
0.19047619047619
|
0.0129032258064516
|
|
GO:BP
|
GO:0009966
|
regulation of signal transduction
|
0.00246385347891222
|
3165
|
69
|
24
|
0.347826086956522
|
0.00758293838862559
|
|
GO:BP
|
GO:0032501
|
multicellular organismal process
|
0.00253767421162414
|
7946
|
57
|
38
|
0.666666666666667
|
0.00478228039265039
|
|
GO:BP
|
GO:1901990
|
regulation of mitotic cell cycle phase transition
|
0.00257146480734839
|
451
|
40
|
6
|
0.15
|
0.0133037694013304
|
|
GO:BP
|
GO:0019222
|
regulation of metabolic process
|
0.0026151545738738
|
7148
|
24
|
18
|
0.75
|
0.00251818690542809
|
|
GO:BP
|
GO:0050900
|
leukocyte migration
|
0.00264600464092257
|
511
|
135
|
12
|
0.0888888888888889
|
0.0234833659491194
|
|
GO:BP
|
GO:0006887
|
exocytosis
|
0.00268910126845822
|
904
|
47
|
9
|
0.191489361702128
|
0.00995575221238938
|
|
GO:BP
|
GO:0055080
|
cation homeostasis
|
0.00282110478790691
|
750
|
135
|
15
|
0.111111111111111
|
0.02
|
|
GO:BP
|
GO:0070647
|
protein modification by small protein conjugation or removal
|
0.00286408204509985
|
1184
|
23
|
7
|
0.304347826086957
|
0.00591216216216216
|
|
GO:BP
|
GO:0046700
|
heterocycle catabolic process
|
0.00290703029053499
|
602
|
56
|
8
|
0.142857142857143
|
0.0132890365448505
|
|
GO:BP
|
GO:0030003
|
cellular cation homeostasis
|
0.00294321584641934
|
673
|
135
|
14
|
0.103703703703704
|
0.0208023774145617
|
|
GO:BP
|
GO:0051336
|
regulation of hydrolase activity
|
0.00294321584641934
|
1321
|
140
|
22
|
0.157142857142857
|
0.0166540499621499
|
|
GO:BP
|
GO:0030888
|
regulation of B cell proliferation
|
0.00305342012894883
|
63
|
107
|
4
|
0.0373831775700935
|
0.0634920634920635
|
|
GO:BP
|
GO:0045930
|
negative regulation of mitotic cell cycle
|
0.00305342012894883
|
333
|
21
|
4
|
0.19047619047619
|
0.012012012012012
|
|
GO:BP
|
GO:0044270
|
cellular nitrogen compound catabolic process
|
0.00306465996906395
|
608
|
56
|
8
|
0.142857142857143
|
0.0131578947368421
|
|
GO:BP
|
GO:0098771
|
inorganic ion homeostasis
|
0.00320420622397674
|
761
|
135
|
15
|
0.111111111111111
|
0.0197109067017083
|
|
GO:BP
|
GO:0048729
|
tissue morphogenesis
|
0.00331475001795532
|
667
|
40
|
7
|
0.175
|
0.0104947526236882
|
|
GO:BP
|
GO:0009628
|
response to abiotic stimulus
|
0.00342215196630016
|
1261
|
43
|
10
|
0.232558139534884
|
0.00793021411578113
|
|
GO:BP
|
GO:0060070
|
canonical Wnt signaling pathway
|
0.00344350645786874
|
345
|
21
|
4
|
0.19047619047619
|
0.0115942028985507
|
|
GO:BP
|
GO:0006873
|
cellular ion homeostasis
|
0.00352121767567089
|
687
|
135
|
14
|
0.103703703703704
|
0.0203784570596798
|
|
GO:BP
|
GO:0019439
|
aromatic compound catabolic process
|
0.00352151095983744
|
623
|
56
|
8
|
0.142857142857143
|
0.0128410914927769
|
|
GO:BP
|
GO:0001959
|
regulation of cytokine-mediated signaling pathway
|
0.00361257654666356
|
184
|
68
|
5
|
0.0735294117647059
|
0.0271739130434783
|
|
GO:BP
|
GO:0072677
|
eosinophil migration
|
0.00364455625706243
|
23
|
135
|
3
|
0.0222222222222222
|
0.130434782608696
|
|
GO:BP
|
GO:0010524
|
positive regulation of calcium ion transport into cytosol
|
0.00370508362311655
|
56
|
55
|
3
|
0.0545454545454545
|
0.0535714285714286
|
|
GO:BP
|
GO:0050789
|
regulation of biological process
|
0.00371177317492733
|
12091
|
58
|
50
|
0.862068965517241
|
0.00413530725332892
|
|
GO:BP
|
GO:0006874
|
cellular calcium ion homeostasis
|
0.00371177317492733
|
460
|
135
|
11
|
0.0814814814814815
|
0.0239130434782609
|
|
GO:BP
|
GO:0070227
|
lymphocyte apoptotic process
|
0.00372709872895474
|
74
|
42
|
3
|
0.0714285714285714
|
0.0405405405405405
|
|
GO:BP
|
GO:1903311
|
regulation of mRNA metabolic process
|
0.00372960821793254
|
356
|
21
|
4
|
0.19047619047619
|
0.0112359550561798
|
|
GO:BP
|
GO:0009892
|
negative regulation of metabolic process
|
0.00374270451442959
|
3479
|
69
|
25
|
0.36231884057971
|
0.00718597298074159
|
|
GO:BP
|
GO:0032753
|
positive regulation of interleukin-4 production
|
0.00376785407924117
|
24
|
132
|
3
|
0.0227272727272727
|
0.125
|
|
GO:BP
|
GO:1901987
|
regulation of cell cycle phase transition
|
0.00378801826743999
|
493
|
40
|
6
|
0.15
|
0.0121703853955375
|
|
GO:BP
|
GO:0051924
|
regulation of calcium ion transport
|
0.00398655083958239
|
259
|
135
|
8
|
0.0592592592592593
|
0.0308880308880309
|
|
GO:BP
|
GO:0001666
|
response to hypoxia
|
0.00412769281542616
|
367
|
21
|
4
|
0.19047619047619
|
0.0108991825613079
|
|
GO:BP
|
GO:0071222
|
cellular response to lipopolysaccharide
|
0.00413483781952902
|
200
|
135
|
7
|
0.0518518518518519
|
0.035
|
|
GO:BP
|
GO:0055085
|
transmembrane transport
|
0.00420310546086966
|
1635
|
41
|
11
|
0.268292682926829
|
0.00672782874617737
|
|
GO:BP
|
GO:0010948
|
negative regulation of cell cycle process
|
0.00427647713294846
|
372
|
21
|
4
|
0.19047619047619
|
0.010752688172043
|
|
GO:BP
|
GO:0090025
|
regulation of monocyte chemotaxis
|
0.00427647713294846
|
27
|
123
|
3
|
0.024390243902439
|
0.111111111111111
|
|
GO:BP
|
GO:0065003
|
protein-containing complex assembly
|
0.00428216811443516
|
1680
|
3
|
3
|
1
|
0.00178571428571429
|
|
GO:BP
|
GO:0032479
|
regulation of type I interferon production
|
0.00431950358461696
|
130
|
101
|
5
|
0.0495049504950495
|
0.0384615384615385
|
|
GO:BP
|
GO:0030111
|
regulation of Wnt signaling pathway
|
0.00438759902737442
|
376
|
21
|
4
|
0.19047619047619
|
0.0106382978723404
|
|
GO:BP
|
GO:1901361
|
organic cyclic compound catabolic process
|
0.00438759902737442
|
651
|
56
|
8
|
0.142857142857143
|
0.0122887864823349
|
|
GO:BP
|
GO:0055074
|
calcium ion homeostasis
|
0.00438759902737442
|
472
|
135
|
11
|
0.0814814814814815
|
0.0233050847457627
|
|
GO:BP
|
GO:0032606
|
type I interferon production
|
0.00440521737664617
|
131
|
101
|
5
|
0.0495049504950495
|
0.0381679389312977
|
|
GO:BP
|
GO:0019752
|
carboxylic acid metabolic process
|
0.00441906881699912
|
1024
|
21
|
6
|
0.285714285714286
|
0.005859375
|
|
GO:BP
|
GO:0060759
|
regulation of response to cytokine stimulus
|
0.00455209939462565
|
197
|
68
|
5
|
0.0735294117647059
|
0.0253807106598985
|
|
GO:BP
|
GO:0001961
|
positive regulation of cytokine-mediated signaling pathway
|
0.00459228033678948
|
50
|
68
|
3
|
0.0441176470588235
|
0.06
|
|
GO:BP
|
GO:0036293
|
response to decreased oxygen levels
|
0.00459840433637363
|
382
|
21
|
4
|
0.19047619047619
|
0.0104712041884817
|
|
GO:BP
|
GO:0045862
|
positive regulation of proteolysis
|
0.00460852138489417
|
378
|
54
|
6
|
0.111111111111111
|
0.0158730158730159
|
|
GO:BP
|
GO:0050920
|
regulation of chemotaxis
|
0.00464879487694575
|
226
|
123
|
7
|
0.0569105691056911
|
0.0309734513274336
|
|
GO:BP
|
GO:0006402
|
mRNA catabolic process
|
0.00464879487694575
|
384
|
21
|
4
|
0.19047619047619
|
0.0104166666666667
|
|
GO:BP
|
GO:0009893
|
positive regulation of metabolic process
|
0.00464879487694575
|
3783
|
32
|
15
|
0.46875
|
0.00396510705789056
|
|
GO:BP
|
GO:0010646
|
regulation of cell communication
|
0.00464879487694575
|
3543
|
69
|
25
|
0.36231884057971
|
0.00705616709003669
|
|
GO:BP
|
GO:2000106
|
regulation of leukocyte apoptotic process
|
0.00468675638574634
|
82
|
42
|
3
|
0.0714285714285714
|
0.0365853658536585
|
|
GO:BP
|
GO:0010959
|
regulation of metal ion transport
|
0.00470212113501375
|
268
|
135
|
8
|
0.0592592592592593
|
0.0298507462686567
|
|
GO:BP
|
GO:0043312
|
neutrophil degranulation
|
0.0048664981003147
|
482
|
8
|
3
|
0.375
|
0.00622406639004149
|
|
GO:BP
|
GO:0002283
|
neutrophil activation involved in immune response
|
0.00493927078830945
|
485
|
8
|
3
|
0.375
|
0.00618556701030928
|
|
GO:BP
|
GO:2000108
|
positive regulation of leukocyte apoptotic process
|
0.0049846125829401
|
29
|
123
|
3
|
0.024390243902439
|
0.103448275862069
|
|
GO:BP
|
GO:0032496
|
response to lipopolysaccharide
|
0.00498473599387022
|
338
|
135
|
9
|
0.0666666666666667
|
0.0266272189349112
|
|
GO:BP
|
GO:0043436
|
oxoacid metabolic process
|
0.00500646245848429
|
1057
|
21
|
6
|
0.285714285714286
|
0.00567644276253548
|
|
GO:BP
|
GO:0051851
|
modulation by host of symbiont process
|
0.00500887683495346
|
65
|
123
|
4
|
0.032520325203252
|
0.0615384615384615
|
|
GO:BP
|
GO:0023051
|
regulation of signaling
|
0.00520600922287995
|
3578
|
69
|
25
|
0.36231884057971
|
0.00698714365567356
|
|
GO:BP
|
GO:0002446
|
neutrophil mediated immunity
|
0.00520600922287995
|
497
|
8
|
3
|
0.375
|
0.00603621730382294
|
|
GO:BP
|
GO:0042119
|
neutrophil activation
|
0.00522262173768473
|
498
|
8
|
3
|
0.375
|
0.00602409638554217
|
|
GO:BP
|
GO:0042742
|
defense response to bacterium
|
0.00522262173768473
|
349
|
132
|
9
|
0.0681818181818182
|
0.0257879656160458
|
|
GO:BP
|
GO:0051649
|
establishment of localization in cell
|
0.00535449736179057
|
2813
|
19
|
9
|
0.473684210526316
|
0.00319943121222894
|
|
GO:BP
|
GO:0051345
|
positive regulation of hydrolase activity
|
0.00535449736179057
|
783
|
140
|
15
|
0.107142857142857
|
0.0191570881226054
|
|
GO:BP
|
GO:0036230
|
granulocyte activation
|
0.00535449736179057
|
504
|
8
|
3
|
0.375
|
0.00595238095238095
|
|
GO:BP
|
GO:1904064
|
positive regulation of cation transmembrane transport
|
0.0054815312655824
|
150
|
55
|
4
|
0.0727272727272727
|
0.0266666666666667
|
|
GO:BP
|
GO:0070482
|
response to oxygen levels
|
0.00552283741575752
|
408
|
21
|
4
|
0.19047619047619
|
0.00980392156862745
|
|
GO:BP
|
GO:0071219
|
cellular response to molecule of bacterial origin
|
0.00553639474267468
|
214
|
135
|
7
|
0.0518518518518519
|
0.0327102803738318
|
|
GO:BP
|
GO:2001056
|
positive regulation of cysteine-type endopeptidase activity
|
0.00561379404153556
|
154
|
54
|
4
|
0.0740740740740741
|
0.025974025974026
|
|
GO:BP
|
GO:0006082
|
organic acid metabolic process
|
0.00569097127017604
|
1090
|
21
|
6
|
0.285714285714286
|
0.0055045871559633
|
|
GO:BP
|
GO:0010522
|
regulation of calcium ion transport into cytosol
|
0.00578542869681826
|
106
|
135
|
5
|
0.037037037037037
|
0.0471698113207547
|
|
GO:BP
|
GO:0070229
|
negative regulation of lymphocyte apoptotic process
|
0.00581389040791658
|
31
|
123
|
3
|
0.024390243902439
|
0.0967741935483871
|
|
GO:BP
|
GO:0007259
|
receptor signaling pathway via JAK-STAT
|
0.00587472718620254
|
174
|
123
|
6
|
0.0487804878048781
|
0.0344827586206897
|
|
GO:BP
|
GO:0072503
|
cellular divalent inorganic cation homeostasis
|
0.00600720216225672
|
497
|
135
|
11
|
0.0814814814814815
|
0.0221327967806841
|
|
GO:BP
|
GO:0002920
|
regulation of humoral immune response
|
0.00603458690051191
|
133
|
109
|
5
|
0.0458715596330275
|
0.037593984962406
|
|
GO:BP
|
GO:0043933
|
protein-containing complex subunit organization
|
0.00608163614572181
|
1963
|
3
|
3
|
1
|
0.00152827305145186
|
|
GO:BP
|
GO:0060760
|
positive regulation of response to cytokine stimulus
|
0.00611943961756043
|
57
|
68
|
3
|
0.0441176470588235
|
0.0526315789473684
|
|
GO:BP
|
GO:0006401
|
RNA catabolic process
|
0.00616012742159498
|
424
|
21
|
4
|
0.19047619047619
|
0.00943396226415094
|
|
GO:BP
|
GO:0002221
|
pattern recognition receptor signaling pathway
|
0.00621289131561901
|
213
|
69
|
5
|
0.072463768115942
|
0.0234741784037559
|
|
GO:BP
|
GO:0048513
|
animal organ development
|
0.00621289131561901
|
3672
|
8
|
6
|
0.75
|
0.00163398692810458
|
|
GO:BP
|
GO:0042531
|
positive regulation of tyrosine phosphorylation of STAT protein
|
0.00648111193842602
|
71
|
123
|
4
|
0.032520325203252
|
0.0563380281690141
|
|
GO:BP
|
GO:0045596
|
negative regulation of cell differentiation
|
0.00674859920646953
|
697
|
33
|
6
|
0.181818181818182
|
0.00860832137733142
|
|
GO:BP
|
GO:0044265
|
cellular macromolecule catabolic process
|
0.00675144098446224
|
1231
|
41
|
9
|
0.219512195121951
|
0.00731112916328188
|
|
GO:BP
|
GO:1904427
|
positive regulation of calcium ion transmembrane transport
|
0.0068608506087211
|
74
|
55
|
3
|
0.0545454545454545
|
0.0405405405405405
|
|
GO:BP
|
GO:0055082
|
cellular chemical homeostasis
|
0.00703029375319539
|
841
|
135
|
15
|
0.111111111111111
|
0.0178359096313912
|
|
GO:BP
|
GO:0097696
|
receptor signaling pathway via STAT
|
0.00741340420002229
|
184
|
123
|
6
|
0.0487804878048781
|
0.0326086956521739
|
|
GO:BP
|
GO:0006810
|
transport
|
0.00747398679255156
|
5224
|
21
|
13
|
0.619047619047619
|
0.0024885145482389
|
|
GO:BP
|
GO:0072507
|
divalent inorganic cation homeostasis
|
0.00762203082949121
|
516
|
135
|
11
|
0.0814814814814815
|
0.0213178294573643
|
|
GO:BP
|
GO:0034655
|
nucleobase-containing compound catabolic process
|
0.00786292348022655
|
567
|
56
|
7
|
0.125
|
0.0123456790123457
|
|
GO:BP
|
GO:1902533
|
positive regulation of intracellular signal transduction
|
0.00788075302888177
|
1037
|
123
|
16
|
0.130081300813008
|
0.0154291224686596
|
|
GO:BP
|
GO:0032940
|
secretion by cell
|
0.00788657506600829
|
1364
|
8
|
4
|
0.5
|
0.00293255131964809
|
|
GO:BP
|
GO:0070232
|
regulation of T cell apoptotic process
|
0.00827375453616005
|
36
|
123
|
3
|
0.024390243902439
|
0.0833333333333333
|
|
GO:BP
|
GO:0071887
|
leukocyte apoptotic process
|
0.00876708412027933
|
108
|
42
|
3
|
0.0714285714285714
|
0.0277777777777778
|
|
GO:BP
|
GO:0140352
|
export from cell
|
0.00887263085420923
|
1418
|
8
|
4
|
0.5
|
0.00282087447108604
|
|
GO:BP
|
GO:0044772
|
mitotic cell cycle phase transition
|
0.0089283968991242
|
609
|
40
|
6
|
0.15
|
0.00985221674876847
|
|
GO:BP
|
GO:0071216
|
cellular response to biotic stimulus
|
0.00900601762318607
|
238
|
135
|
7
|
0.0518518518518519
|
0.0294117647058824
|
|
GO:BP
|
GO:1903169
|
regulation of calcium ion transmembrane transport
|
0.0090834308179629
|
158
|
62
|
4
|
0.0645161290322581
|
0.0253164556962025
|
|
GO:BP
|
GO:0050729
|
positive regulation of inflammatory response
|
0.00910381320402572
|
142
|
69
|
4
|
0.0579710144927536
|
0.028169014084507
|
|
GO:BP
|
GO:0032480
|
negative regulation of type I interferon production
|
0.00924073093611481
|
46
|
101
|
3
|
0.0297029702970297
|
0.0652173913043478
|
|
GO:BP
|
GO:0051234
|
establishment of localization
|
0.00927368594344208
|
5372
|
21
|
13
|
0.619047619047619
|
0.00241995532390171
|
|
GO:BP
|
GO:0051279
|
regulation of release of sequestered calcium ion into cytosol
|
0.00942098621628598
|
85
|
55
|
3
|
0.0545454545454545
|
0.0352941176470588
|
|
GO:BP
|
GO:0007346
|
regulation of mitotic cell cycle
|
0.00948930091384165
|
619
|
40
|
6
|
0.15
|
0.00969305331179321
|
|
GO:BP
|
GO:1901224
|
positive regulation of NIK/NF-kappaB signaling
|
0.00949852828306695
|
68
|
69
|
3
|
0.0434782608695652
|
0.0441176470588235
|
|
GO:BP
|
GO:0050731
|
positive regulation of peptidyl-tyrosine phosphorylation
|
0.0098655076895628
|
198
|
123
|
6
|
0.0487804878048781
|
0.0303030303030303
|
|
GO:BP
|
GO:0002455
|
humoral immune response mediated by circulating immunoglobulin
|
0.0105476424654274
|
156
|
109
|
5
|
0.0458715596330275
|
0.032051282051282
|
|
GO:BP
|
GO:0032612
|
interleukin-1 production
|
0.010549493237439
|
126
|
135
|
5
|
0.037037037037037
|
0.0396825396825397
|
|
GO:BP
|
GO:0042100
|
B cell proliferation
|
0.0111980412741635
|
98
|
107
|
4
|
0.0373831775700935
|
0.0408163265306122
|
|
GO:BP
|
GO:0045786
|
negative regulation of cell cycle
|
0.0113113547040084
|
647
|
40
|
6
|
0.15
|
0.00927357032457496
|
|
GO:BP
|
GO:0035556
|
intracellular signal transduction
|
0.0113113547040084
|
2772
|
69
|
20
|
0.289855072463768
|
0.00721500721500722
|
|
GO:BP
|
GO:0016055
|
Wnt signaling pathway
|
0.011739651845471
|
529
|
21
|
4
|
0.19047619047619
|
0.00756143667296786
|
|
GO:BP
|
GO:0042509
|
regulation of tyrosine phosphorylation of STAT protein
|
0.0117794679774835
|
87
|
123
|
4
|
0.032520325203252
|
0.0459770114942529
|
|
GO:BP
|
GO:1901214
|
regulation of neuron death
|
0.0118013293720897
|
323
|
135
|
8
|
0.0592592592592593
|
0.0247678018575851
|
|
GO:BP
|
GO:0198738
|
cell-cell signaling by wnt
|
0.0118115581491315
|
531
|
21
|
4
|
0.19047619047619
|
0.00753295668549906
|
|
GO:BP
|
GO:0044770
|
cell cycle phase transition
|
0.012009152850609
|
658
|
40
|
6
|
0.15
|
0.00911854103343465
|
|
GO:BP
|
GO:0007204
|
positive regulation of cytosolic calcium ion concentration
|
0.0121321844121026
|
325
|
135
|
8
|
0.0592592592592593
|
0.0246153846153846
|
|
GO:BP
|
GO:0007260
|
tyrosine phosphorylation of STAT protein
|
0.0124751001745619
|
89
|
123
|
4
|
0.032520325203252
|
0.0449438202247191
|
|
GO:BP
|
GO:0019538
|
protein metabolic process
|
0.0129437353371619
|
5808
|
73
|
35
|
0.479452054794521
|
0.00602617079889807
|
|
GO:BP
|
GO:0030154
|
cell differentiation
|
0.0131590024001895
|
4350
|
8
|
6
|
0.75
|
0.00137931034482759
|
|
GO:BP
|
GO:0065007
|
biological regulation
|
0.0141468902605834
|
12809
|
73
|
62
|
0.849315068493151
|
0.00484034663127488
|
|
GO:BP
|
GO:0051050
|
positive regulation of transport
|
0.0142455982875756
|
923
|
135
|
15
|
0.111111111111111
|
0.0162513542795233
|
|
GO:BP
|
GO:0048869
|
cellular developmental process
|
0.0143745891764327
|
4428
|
8
|
6
|
0.75
|
0.0013550135501355
|
|
GO:BP
|
GO:0002820
|
negative regulation of adaptive immune response
|
0.0144733666261292
|
51
|
111
|
3
|
0.027027027027027
|
0.0588235294117647
|
|
GO:BP
|
GO:0051246
|
regulation of protein metabolic process
|
0.0145126120792033
|
2704
|
58
|
17
|
0.293103448275862
|
0.00628698224852071
|
|
GO:BP
|
GO:0051702
|
biological process involved in interaction with symbiont
|
0.0157892440908036
|
96
|
123
|
4
|
0.032520325203252
|
0.0416666666666667
|
|
GO:BP
|
GO:0002381
|
immunoglobulin production involved in immunoglobulin-mediated immune response
|
0.0159285485937589
|
53
|
111
|
3
|
0.027027027027027
|
0.0566037735849057
|
|
GO:BP
|
GO:0034767
|
positive regulation of ion transmembrane transport
|
0.0160002629166726
|
217
|
55
|
4
|
0.0727272727272727
|
0.0184331797235023
|
|
GO:BP
|
GO:0034764
|
positive regulation of transmembrane transport
|
0.0160002629166726
|
217
|
55
|
4
|
0.0727272727272727
|
0.0184331797235023
|
|
GO:BP
|
GO:0030449
|
regulation of complement activation
|
0.016545827012613
|
110
|
109
|
4
|
0.036697247706422
|
0.0363636363636364
|
|
GO:BP
|
GO:0002889
|
regulation of immunoglobulin mediated immune response
|
0.0166343581441941
|
54
|
111
|
3
|
0.027027027027027
|
0.0555555555555556
|
|
GO:BP
|
GO:0002712
|
regulation of B cell mediated immunity
|
0.0166343581441941
|
54
|
111
|
3
|
0.027027027027027
|
0.0555555555555556
|
|
GO:BP
|
GO:0048878
|
chemical homeostasis
|
0.0168039668265225
|
1229
|
135
|
18
|
0.133333333333333
|
0.0146460537021969
|
|
GO:BP
|
GO:0022607
|
cellular component assembly
|
0.0171078061628795
|
3010
|
3
|
3
|
1
|
0.000996677740863787
|
|
GO:BP
|
GO:0006956
|
complement activation
|
0.0172831974918395
|
180
|
109
|
5
|
0.0458715596330275
|
0.0277777777777778
|
|
GO:BP
|
GO:0009057
|
macromolecule catabolic process
|
0.0176828435303492
|
1468
|
41
|
9
|
0.219512195121951
|
0.0061307901907357
|
|
GO:BP
|
GO:0007267
|
cell-cell signaling
|
0.0177191431848729
|
1722
|
55
|
12
|
0.218181818181818
|
0.00696864111498258
|
|
GO:BP
|
GO:0051817
|
modulation of process of other organism involved in symbiotic interaction
|
0.0187476106560729
|
102
|
123
|
4
|
0.032520325203252
|
0.0392156862745098
|
|
GO:BP
|
GO:0051641
|
cellular localization
|
0.018918702583619
|
3541
|
43
|
16
|
0.372093023255814
|
0.00451849759954815
|
|
GO:BP
|
GO:0006968
|
cellular defense response
|
0.0190067191275658
|
53
|
121
|
3
|
0.0247933884297521
|
0.0566037735849057
|
|
GO:BP
|
GO:0050790
|
regulation of catalytic activity
|
0.0196557622865999
|
2545
|
21
|
8
|
0.380952380952381
|
0.0031434184675835
|
|
GO:BP
|
GO:0070231
|
T cell apoptotic process
|
0.0197784623339297
|
53
|
123
|
3
|
0.024390243902439
|
0.0566037735849057
|
|
GO:BP
|
GO:1905114
|
cell surface receptor signaling pathway involved in cell-cell signaling
|
0.0197784623339297
|
636
|
21
|
4
|
0.19047619047619
|
0.00628930817610063
|
|
GO:BP
|
GO:0051480
|
regulation of cytosolic calcium ion concentration
|
0.0198084020938086
|
360
|
135
|
8
|
0.0592592592592593
|
0.0222222222222222
|
|
GO:BP
|
GO:0007584
|
response to nutrient
|
0.0200076017379524
|
182
|
113
|
5
|
0.0442477876106195
|
0.0274725274725275
|
|
GO:BP
|
GO:0051289
|
protein homotetramerization
|
0.0200076017379524
|
56
|
117
|
3
|
0.0256410256410256
|
0.0535714285714286
|
|
GO:BP
|
GO:0044085
|
cellular component biogenesis
|
0.0202450930797048
|
3257
|
3
|
3
|
1
|
0.00092109303039607
|
|
GO:BP
|
GO:0070997
|
neuron death
|
0.0209449492686939
|
365
|
135
|
8
|
0.0592592592592593
|
0.0219178082191781
|
|
GO:BP
|
GO:0032879
|
regulation of localization
|
0.0212206490621535
|
2872
|
135
|
33
|
0.244444444444444
|
0.0114902506963788
|
|
GO:BP
|
GO:0051209
|
release of sequestered calcium ion into cytosol
|
0.0212206490621535
|
123
|
55
|
3
|
0.0545454545454545
|
0.024390243902439
|
|
GO:BP
|
GO:0016445
|
somatic diversification of immunoglobulins
|
0.021221980014858
|
61
|
111
|
3
|
0.027027027027027
|
0.0491803278688525
|
|
GO:BP
|
GO:0051283
|
negative regulation of sequestering of calcium ion
|
0.0215347421002336
|
124
|
55
|
3
|
0.0545454545454545
|
0.0241935483870968
|
|
GO:BP
|
GO:0019725
|
cellular homeostasis
|
0.021727326021155
|
981
|
135
|
15
|
0.111111111111111
|
0.0152905198776758
|
|
GO:BP
|
GO:0065009
|
regulation of molecular function
|
0.0218660672839658
|
3221
|
21
|
9
|
0.428571428571429
|
0.00279416330332195
|
|
GO:BP
|
GO:0051651
|
maintenance of location in cell
|
0.0218660672839658
|
223
|
135
|
6
|
0.0444444444444444
|
0.0269058295964126
|
|
GO:BP
|
GO:0009612
|
response to mechanical stimulus
|
0.0218660672839658
|
218
|
32
|
3
|
0.09375
|
0.0137614678899083
|
|
GO:BP
|
GO:0051282
|
regulation of sequestering of calcium ion
|
0.0218660672839658
|
126
|
55
|
3
|
0.0545454545454545
|
0.0238095238095238
|
|
GO:BP
|
GO:0006915
|
apoptotic process
|
0.0222075544139514
|
1990
|
136
|
25
|
0.183823529411765
|
0.0125628140703518
|
|
GO:BP
|
GO:0050864
|
regulation of B cell activation
|
0.0224791177382755
|
194
|
111
|
5
|
0.045045045045045
|
0.0257731958762887
|
|
GO:BP
|
GO:0032651
|
regulation of interleukin-1 beta production
|
0.0232996419115371
|
102
|
135
|
4
|
0.0296296296296296
|
0.0392156862745098
|
|
GO:BP
|
GO:0051049
|
regulation of transport
|
0.0233709453230461
|
1809
|
42
|
10
|
0.238095238095238
|
0.00552791597567717
|
|
GO:BP
|
GO:0051208
|
sequestering of calcium ion
|
0.0234802788843794
|
130
|
55
|
3
|
0.0545454545454545
|
0.0230769230769231
|
|
GO:BP
|
GO:0048731
|
system development
|
0.0234838063680443
|
5052
|
17
|
10
|
0.588235294117647
|
0.00197941409342835
|
|
GO:BP
|
GO:0051092
|
positive regulation of NF-kappaB transcription factor activity
|
0.0236698820588789
|
162
|
135
|
5
|
0.037037037037037
|
0.0308641975308642
|
|
GO:BP
|
GO:0043269
|
regulation of ion transport
|
0.023801483750225
|
1379
|
6
|
3
|
0.5
|
0.00217548948513416
|
|
GO:BP
|
GO:0033554
|
cellular response to stress
|
0.0243756765991082
|
2111
|
21
|
7
|
0.333333333333333
|
0.00331596399810516
|
|
GO:BP
|
GO:0051093
|
negative regulation of developmental process
|
0.0244476105708336
|
977
|
33
|
6
|
0.181818181818182
|
0.00614124872057318
|
|
GO:BP
|
GO:1904062
|
regulation of cation transmembrane transport
|
0.0245887686999815
|
360
|
62
|
5
|
0.0806451612903226
|
0.0138888888888889
|
|
GO:BP
|
GO:1901564
|
organonitrogen compound metabolic process
|
0.024669010627019
|
6792
|
73
|
38
|
0.520547945205479
|
0.00559481743227326
|
|
GO:BP
|
GO:0050727
|
regulation of inflammatory response
|
0.0247301947683649
|
381
|
135
|
8
|
0.0592592592592593
|
0.020997375328084
|
|
GO:BP
|
GO:0051048
|
negative regulation of secretion
|
0.0248523162147117
|
176
|
42
|
3
|
0.0714285714285714
|
0.0170454545454545
|
|
GO:BP
|
GO:0060402
|
calcium ion transport into cytosol
|
0.0250668823051331
|
165
|
135
|
5
|
0.037037037037037
|
0.0303030303030303
|
|
GO:BP
|
GO:0002637
|
regulation of immunoglobulin production
|
0.0254625060274759
|
67
|
111
|
3
|
0.027027027027027
|
0.0447761194029851
|
|
GO:BP
|
GO:0010564
|
regulation of cell cycle process
|
0.0257221016544795
|
809
|
40
|
6
|
0.15
|
0.00741656365883807
|
|
GO:BP
|
GO:0010941
|
regulation of cell death
|
0.0262661076125354
|
1720
|
135
|
22
|
0.162962962962963
|
0.0127906976744186
|
|
GO:BP
|
GO:0032611
|
interleukin-1 beta production
|
0.0263801601033695
|
107
|
135
|
4
|
0.0296296296296296
|
0.0373831775700935
|
|
GO:BP
|
GO:0070936
|
protein K48-linked ubiquitination
|
0.0278766076676199
|
61
|
127
|
3
|
0.0236220472440945
|
0.0491803278688525
|
|
GO:BP
|
GO:0044093
|
positive regulation of molecular function
|
0.0280639173926944
|
1771
|
140
|
23
|
0.164285714285714
|
0.012987012987013
|
|
GO:BP
|
GO:1901222
|
regulation of NIK/NF-kappaB signaling
|
0.0283417466948765
|
113
|
69
|
3
|
0.0434782608695652
|
0.0265486725663717
|
|
GO:BP
|
GO:0032270
|
positive regulation of cellular protein metabolic process
|
0.0286562833223014
|
1477
|
69
|
12
|
0.173913043478261
|
0.00812457684495599
|
|
GO:BP
|
GO:0097553
|
calcium ion transmembrane import into cytosol
|
0.0296054601543015
|
145
|
55
|
3
|
0.0545454545454545
|
0.0206896551724138
|
|
GO:BP
|
GO:0002200
|
somatic diversification of immune receptors
|
0.0298688074317326
|
72
|
111
|
3
|
0.027027027027027
|
0.0416666666666667
|
|
GO:BP
|
GO:0030335
|
positive regulation of cell migration
|
0.0303063515122151
|
567
|
135
|
10
|
0.0740740740740741
|
0.017636684303351
|
|
GO:BP
|
GO:0050730
|
regulation of peptidyl-tyrosine phosphorylation
|
0.0307061514540175
|
269
|
123
|
6
|
0.0487804878048781
|
0.0223048327137546
|
|
GO:BP
|
GO:0007189
|
adenylate cyclase-activating G protein-coupled receptor signaling pathway
|
0.030978942227136
|
148
|
55
|
3
|
0.0545454545454545
|
0.0202702702702703
|
|
GO:BP
|
GO:2000045
|
regulation of G1/S transition of mitotic cell cycle
|
0.0313917812358276
|
182
|
131
|
5
|
0.0381679389312977
|
0.0274725274725275
|
|
GO:BP
|
GO:0044267
|
cellular protein metabolic process
|
0.0314765627546303
|
5229
|
54
|
24
|
0.444444444444444
|
0.00458978772231784
|
|
GO:BP
|
GO:0006958
|
complement activation, classical pathway
|
0.031749096633504
|
142
|
109
|
4
|
0.036697247706422
|
0.028169014084507
|
|
GO:BP
|
GO:0042981
|
regulation of apoptotic process
|
0.0320658627565933
|
1549
|
135
|
20
|
0.148148148148148
|
0.012911555842479
|
|
GO:BP
|
GO:0010942
|
positive regulation of cell death
|
0.0321423033999012
|
684
|
131
|
11
|
0.083969465648855
|
0.0160818713450292
|
|
GO:BP
|
GO:0008219
|
cell death
|
0.032975350810988
|
2292
|
136
|
27
|
0.198529411764706
|
0.0117801047120419
|
|
GO:BP
|
GO:0035821
|
modulation of process of other organism
|
0.033411025680033
|
128
|
123
|
4
|
0.032520325203252
|
0.03125
|
|
GO:BP
|
GO:0043170
|
macromolecule metabolic process
|
0.0334819986425521
|
9796
|
59
|
41
|
0.694915254237288
|
0.0041853817884851
|
|
GO:BP
|
GO:0007568
|
aging
|
0.0341582478787277
|
329
|
135
|
7
|
0.0518518518518519
|
0.0212765957446809
|
|
GO:BP
|
GO:0009887
|
animal organ morphogenesis
|
0.0346458826399173
|
1073
|
24
|
5
|
0.208333333333333
|
0.00465983224603914
|
|
GO:BP
|
GO:0032652
|
regulation of interleukin-1 production
|
0.0350650970818596
|
119
|
135
|
4
|
0.0296296296296296
|
0.0336134453781513
|
|
GO:BP
|
GO:0032268
|
regulation of cellular protein metabolic process
|
0.0351073551551972
|
2541
|
58
|
15
|
0.258620689655172
|
0.00590318772136954
|
|
GO:BP
|
GO:0051051
|
negative regulation of transport
|
0.0362220745310626
|
474
|
19
|
3
|
0.157894736842105
|
0.00632911392405063
|
|
GO:BP
|
GO:0007275
|
multicellular organism development
|
0.036383949217453
|
5613
|
6
|
5
|
0.833333333333333
|
0.00089078923926599
|
|
GO:BP
|
GO:0060401
|
cytosolic calcium ion transport
|
0.0367072533197327
|
186
|
135
|
5
|
0.037037037037037
|
0.0268817204301075
|
|
GO:BP
|
GO:0050766
|
positive regulation of phagocytosis
|
0.0368526470376699
|
67
|
132
|
3
|
0.0227272727272727
|
0.0447761194029851
|
|
GO:BP
|
GO:0043085
|
positive regulation of catalytic activity
|
0.0369439362183969
|
1418
|
140
|
19
|
0.135714285714286
|
0.0133991537376587
|
|
GO:BP
|
GO:0060341
|
regulation of cellular localization
|
0.037089852689681
|
851
|
42
|
6
|
0.142857142857143
|
0.00705052878965922
|
|
GO:BP
|
GO:2000147
|
positive regulation of cell motility
|
0.0371329975540264
|
591
|
135
|
10
|
0.0740740740740741
|
0.0169204737732657
|
|
GO:BP
|
GO:0043067
|
regulation of programmed cell death
|
0.0378489489357415
|
1582
|
135
|
20
|
0.148148148148148
|
0.0126422250316056
|
|
GO:BP
|
GO:1903047
|
mitotic cell cycle process
|
0.0396243746133969
|
911
|
40
|
6
|
0.15
|
0.00658616904500549
|
|
GO:BP
|
GO:0050918
|
positive chemotaxis
|
0.0399443794338606
|
68
|
135
|
3
|
0.0222222222222222
|
0.0441176470588235
|
|
GO:BP
|
GO:0051247
|
positive regulation of protein metabolic process
|
0.0401087780107602
|
1564
|
69
|
12
|
0.173913043478261
|
0.00767263427109974
|
|
GO:BP
|
GO:0051272
|
positive regulation of cellular component movement
|
0.0416235493140779
|
604
|
135
|
10
|
0.0740740740740741
|
0.0165562913907285
|
|
GO:BP
|
GO:0043547
|
positive regulation of GTPase activity
|
0.0416235493140779
|
413
|
140
|
8
|
0.0571428571428571
|
0.0193704600484262
|
|
GO:BP
|
GO:0040017
|
positive regulation of locomotion
|
0.0423204638035385
|
606
|
135
|
10
|
0.0740740740740741
|
0.0165016501650165
|
|
GO:BP
|
GO:0042113
|
B cell activation
|
0.0438427950632514
|
330
|
111
|
6
|
0.0540540540540541
|
0.0181818181818182
|
|
GO:BP
|
GO:0031329
|
regulation of cellular catabolic process
|
0.0442457443326039
|
866
|
21
|
4
|
0.19047619047619
|
0.0046189376443418
|
|
GO:BP
|
GO:0010812
|
negative regulation of cell-substrate adhesion
|
0.0445440099389398
|
69
|
140
|
3
|
0.0214285714285714
|
0.0434782608695652
|
|
GO:BP
|
GO:0012501
|
programmed cell death
|
0.0446789567033564
|
2143
|
136
|
25
|
0.183823529411765
|
0.0116658889407373
|
|
GO:BP
|
GO:0006816
|
calcium ion transport
|
0.0449396738974401
|
436
|
135
|
8
|
0.0592592592592593
|
0.018348623853211
|
|
GO:BP
|
GO:0060627
|
regulation of vesicle-mediated transport
|
0.0452816911188616
|
536
|
132
|
9
|
0.0681818181818182
|
0.0167910447761194
|
|
GO:BP
|
GO:0016071
|
mRNA metabolic process
|
0.0453213149716658
|
874
|
21
|
4
|
0.19047619047619
|
0.0045766590389016
|
|
GO:BP
|
GO:0030334
|
regulation of cell migration
|
0.0457849111533855
|
999
|
135
|
14
|
0.103703703703704
|
0.014014014014014
|
|
GO:BP
|
GO:1902806
|
regulation of cell cycle G1/S phase transition
|
0.0458463740211678
|
206
|
131
|
5
|
0.0381679389312977
|
0.0242718446601942
|
|
GO:BP
|
GO:0051726
|
regulation of cell cycle
|
0.0465576745272205
|
1236
|
40
|
7
|
0.175
|
0.00566343042071197
|
|
GO:BP
|
GO:0045765
|
regulation of angiogenesis
|
0.0475358044524769
|
358
|
135
|
7
|
0.0518518518518519
|
0.0195530726256983
|
|
GO:BP
|
GO:0010608
|
posttranscriptional regulation of gene expression
|
0.0481052919273091
|
896
|
21
|
4
|
0.19047619047619
|
0.00446428571428571
|
|
GO:BP
|
GO:0001960
|
negative regulation of cytokine-mediated signaling pathway
|
0.0484150294535493
|
82
|
123
|
3
|
0.024390243902439
|
0.0365853658536585
|
|
GO:BP
|
GO:0048856
|
anatomical structure development
|
0.0491886706090897
|
6106
|
6
|
5
|
0.833333333333333
|
0.000818866688503112
|
|
GO:BP
|
GO:1901575
|
organic substance catabolic process
|
0.0493261228285778
|
2267
|
56
|
13
|
0.232142857142857
|
0.00573445081605646
|
|
GO:BP
|
GO:0044281
|
small molecule metabolic process
|
0.0493261228285778
|
1921
|
21
|
6
|
0.285714285714286
|
0.00312337324310255
|
|
GO:BP
|
GO:0006464
|
cellular protein modification process
|
0.0499314265099958
|
4041
|
23
|
10
|
0.434782608695652
|
0.00247463499133878
|
|
GO:BP
|
GO:0036211
|
protein modification process
|
0.0499314265099958
|
4041
|
23
|
10
|
0.434782608695652
|
0.00247463499133878
|
|
GO:CC
|
GO:0042611
|
MHC protein complex
|
1.20961862310406e-37
|
24
|
25
|
15
|
0.6
|
0.625
|
|
GO:CC
|
GO:0071556
|
integral component of lumenal side of endoplasmic reticulum membrane
|
2.22870061642121e-25
|
25
|
23
|
11
|
0.478260869565217
|
0.44
|
|
GO:CC
|
GO:0098553
|
lumenal side of endoplasmic reticulum membrane
|
2.22870061642121e-25
|
25
|
23
|
11
|
0.478260869565217
|
0.44
|
|
GO:CC
|
GO:0098576
|
lumenal side of membrane
|
7.22396640889448e-24
|
33
|
23
|
11
|
0.478260869565217
|
0.333333333333333
|
|
GO:CC
|
GO:0012507
|
ER to Golgi transport vesicle membrane
|
1.32238062817404e-23
|
57
|
23
|
12
|
0.521739130434783
|
0.210526315789474
|
|
GO:CC
|
GO:0042613
|
MHC class II protein complex
|
1.41480489497146e-21
|
16
|
25
|
9
|
0.36
|
0.5625
|
|
GO:CC
|
GO:0030134
|
COPII-coated ER to Golgi transport vesicle
|
3.59350450190215e-21
|
90
|
23
|
12
|
0.521739130434783
|
0.133333333333333
|
|
GO:CC
|
GO:0030176
|
integral component of endoplasmic reticulum membrane
|
2.31544966533442e-20
|
158
|
23
|
13
|
0.565217391304348
|
0.0822784810126582
|
|
GO:CC
|
GO:0030666
|
endocytic vesicle membrane
|
2.31544966533442e-20
|
159
|
23
|
13
|
0.565217391304348
|
0.0817610062893082
|
|
GO:CC
|
GO:0031227
|
intrinsic component of endoplasmic reticulum membrane
|
3.71614758411939e-20
|
166
|
23
|
13
|
0.565217391304348
|
0.0783132530120482
|
|
GO:CC
|
GO:0030662
|
coated vesicle membrane
|
1.07410312934012e-17
|
177
|
23
|
12
|
0.521739130434783
|
0.0677966101694915
|
|
GO:CC
|
GO:0030658
|
transport vesicle membrane
|
7.5449364813774e-17
|
209
|
23
|
12
|
0.521739130434783
|
0.0574162679425837
|
|
GO:CC
|
GO:0030139
|
endocytic vesicle
|
9.46570591826451e-17
|
306
|
23
|
13
|
0.565217391304348
|
0.042483660130719
|
|
GO:CC
|
GO:0010008
|
endosome membrane
|
5.77537358014492e-16
|
505
|
27
|
15
|
0.555555555555556
|
0.0297029702970297
|
|
GO:CC
|
GO:0042612
|
MHC class I protein complex
|
9.28037250560117e-16
|
8
|
23
|
6
|
0.260869565217391
|
0.75
|
|
GO:CC
|
GO:0031301
|
integral component of organelle membrane
|
1.39498000132143e-15
|
382
|
23
|
13
|
0.565217391304348
|
0.0340314136125654
|
|
GO:CC
|
GO:0031300
|
intrinsic component of organelle membrane
|
3.41996395852535e-15
|
413
|
23
|
13
|
0.565217391304348
|
0.0314769975786925
|
|
GO:CC
|
GO:0030135
|
coated vesicle
|
3.41996395852535e-15
|
295
|
23
|
12
|
0.521739130434783
|
0.0406779661016949
|
|
GO:CC
|
GO:0098797
|
plasma membrane protein complex
|
9.52217499799404e-15
|
694
|
25
|
15
|
0.6
|
0.0216138328530259
|
|
GO:CC
|
GO:0005768
|
endosome
|
1.08506362437512e-14
|
987
|
27
|
17
|
0.62962962962963
|
0.0172239108409321
|
|
GO:CC
|
GO:0030133
|
transport vesicle
|
1.65321226866326e-13
|
414
|
23
|
12
|
0.521739130434783
|
0.0289855072463768
|
|
GO:CC
|
GO:0030659
|
cytoplasmic vesicle membrane
|
2.9181040720276e-13
|
779
|
23
|
14
|
0.608695652173913
|
0.0179717586649551
|
|
GO:CC
|
GO:0012506
|
vesicle membrane
|
4.02419853577868e-13
|
800
|
23
|
14
|
0.608695652173913
|
0.0175
|
|
GO:CC
|
GO:0005765
|
lysosomal membrane
|
6.54795781635543e-13
|
383
|
27
|
12
|
0.444444444444444
|
0.031331592689295
|
|
GO:CC
|
GO:0098852
|
lytic vacuole membrane
|
6.54795781635543e-13
|
383
|
27
|
12
|
0.444444444444444
|
0.031331592689295
|
|
GO:CC
|
GO:0098796
|
membrane protein complex
|
9.8006432114444e-13
|
1322
|
15
|
13
|
0.866666666666667
|
0.00983358547655068
|
|
GO:CC
|
GO:0005774
|
vacuolar membrane
|
3.22733338738953e-12
|
441
|
27
|
12
|
0.444444444444444
|
0.0272108843537415
|
|
GO:CC
|
GO:0031982
|
vesicle
|
7.41263937484634e-12
|
4055
|
28
|
24
|
0.857142857142857
|
0.00591861898890259
|
|
GO:CC
|
GO:0098588
|
bounding membrane of organelle
|
1.07136675968475e-11
|
2158
|
27
|
19
|
0.703703703703704
|
0.00880444856348471
|
|
GO:CC
|
GO:0098552
|
side of membrane
|
1.31798953028586e-11
|
619
|
23
|
12
|
0.521739130434783
|
0.0193861066235864
|
|
GO:CC
|
GO:0030670
|
phagocytic vesicle membrane
|
1.31798953028586e-11
|
76
|
36
|
8
|
0.222222222222222
|
0.105263157894737
|
|
GO:CC
|
GO:0009986
|
cell surface
|
2.13673176762657e-11
|
899
|
97
|
25
|
0.257731958762887
|
0.0278086763070078
|
|
GO:CC
|
GO:0030669
|
clathrin-coated endocytic vesicle membrane
|
2.13673176762657e-11
|
36
|
22
|
6
|
0.272727272727273
|
0.166666666666667
|
|
GO:CC
|
GO:0005789
|
endoplasmic reticulum membrane
|
2.59810727393766e-11
|
1117
|
23
|
14
|
0.608695652173913
|
0.0125335720680394
|
|
GO:CC
|
GO:0098827
|
endoplasmic reticulum subcompartment
|
2.68142911846673e-11
|
1122
|
23
|
14
|
0.608695652173913
|
0.0124777183600713
|
|
GO:CC
|
GO:0042175
|
nuclear outer membrane-endoplasmic reticulum membrane network
|
3.19622348712424e-11
|
1139
|
23
|
14
|
0.608695652173913
|
0.0122914837576822
|
|
GO:CC
|
GO:0045335
|
phagocytic vesicle
|
1.0190803360146e-10
|
137
|
41
|
9
|
0.219512195121951
|
0.0656934306569343
|
|
GO:CC
|
GO:0098791
|
Golgi apparatus subcompartment
|
1.09566269075383e-10
|
885
|
35
|
15
|
0.428571428571429
|
0.0169491525423729
|
|
GO:CC
|
GO:0000139
|
Golgi membrane
|
1.3717765965683e-10
|
771
|
23
|
12
|
0.521739130434783
|
0.0155642023346304
|
|
GO:CC
|
GO:0045334
|
clathrin-coated endocytic vesicle
|
1.53901205932121e-10
|
54
|
9
|
5
|
0.555555555555556
|
0.0925925925925926
|
|
GO:CC
|
GO:0000323
|
lytic vacuole
|
5.62728924465476e-10
|
712
|
27
|
12
|
0.444444444444444
|
0.0168539325842697
|
|
GO:CC
|
GO:0005764
|
lysosome
|
5.62728924465476e-10
|
712
|
27
|
12
|
0.444444444444444
|
0.0168539325842697
|
|
GO:CC
|
GO:0032991
|
protein-containing complex
|
5.91772743576604e-10
|
5548
|
25
|
23
|
0.92
|
0.00414563806777217
|
|
GO:CC
|
GO:0031410
|
cytoplasmic vesicle
|
8.43272325215623e-10
|
2424
|
27
|
18
|
0.666666666666667
|
0.00742574257425743
|
|
GO:CC
|
GO:0097708
|
intracellular vesicle
|
8.47728113422559e-10
|
2428
|
27
|
18
|
0.666666666666667
|
0.00741350906095552
|
|
GO:CC
|
GO:0032588
|
trans-Golgi network membrane
|
2.25740725372387e-09
|
94
|
9
|
5
|
0.555555555555556
|
0.0531914893617021
|
|
GO:CC
|
GO:0005773
|
vacuole
|
2.25740725372387e-09
|
810
|
27
|
12
|
0.444444444444444
|
0.0148148148148148
|
|
GO:CC
|
GO:0031984
|
organelle subcompartment
|
3.19096310330596e-09
|
1773
|
36
|
18
|
0.5
|
0.0101522842639594
|
|
GO:CC
|
GO:0055038
|
recycling endosome membrane
|
3.88981418993989e-09
|
84
|
23
|
6
|
0.260869565217391
|
0.0714285714285714
|
|
GO:CC
|
GO:0030665
|
clathrin-coated vesicle membrane
|
5.65507746479904e-09
|
114
|
9
|
5
|
0.555555555555556
|
0.043859649122807
|
|
GO:CC
|
GO:0042824
|
MHC class I peptide loading complex
|
8.53965899590558e-09
|
9
|
36
|
4
|
0.111111111111111
|
0.444444444444444
|
|
GO:CC
|
GO:0070062
|
extracellular exosome
|
9.18950514316339e-09
|
2176
|
23
|
15
|
0.652173913043478
|
0.00689338235294118
|
|
GO:CC
|
GO:0043230
|
extracellular organelle
|
1.535921240294e-08
|
2263
|
23
|
15
|
0.652173913043478
|
0.0066283694211224
|
|
GO:CC
|
GO:1903561
|
extracellular vesicle
|
1.535921240294e-08
|
2261
|
23
|
15
|
0.652173913043478
|
0.00663423264042459
|
|
GO:CC
|
GO:0005770
|
late endosome
|
1.78863726333209e-08
|
275
|
27
|
8
|
0.296296296296296
|
0.0290909090909091
|
|
GO:CC
|
GO:0005783
|
endoplasmic reticulum
|
3.31535893612869e-08
|
1982
|
23
|
14
|
0.608695652173913
|
0.0070635721493441
|
|
GO:CC
|
GO:0005794
|
Golgi apparatus
|
3.56310299036793e-08
|
1623
|
23
|
13
|
0.565217391304348
|
0.00800985828712261
|
|
GO:CC
|
GO:0031090
|
organelle membrane
|
4.98870608345064e-08
|
3612
|
27
|
19
|
0.703703703703704
|
0.00526024363233666
|
|
GO:CC
|
GO:0030136
|
clathrin-coated vesicle
|
5.61054080968111e-08
|
193
|
4
|
4
|
1
|
0.0207253886010363
|
|
GO:CC
|
GO:0031901
|
early endosome membrane
|
1.50450759517292e-07
|
159
|
23
|
6
|
0.260869565217391
|
0.0377358490566038
|
|
GO:CC
|
GO:0005802
|
trans-Golgi network
|
1.53875467796556e-07
|
250
|
4
|
4
|
1
|
0.016
|
|
GO:CC
|
GO:0031902
|
late endosome membrane
|
1.94483029491727e-07
|
140
|
27
|
6
|
0.222222222222222
|
0.0428571428571429
|
|
GO:CC
|
GO:0005615
|
extracellular space
|
2.95411376287309e-07
|
3594
|
123
|
49
|
0.398373983739837
|
0.0136338341680579
|
|
GO:CC
|
GO:0009897
|
external side of plasma membrane
|
3.58873924892197e-07
|
408
|
107
|
14
|
0.130841121495327
|
0.0343137254901961
|
|
GO:CC
|
GO:0055037
|
recycling endosome
|
3.76634250056062e-07
|
188
|
23
|
6
|
0.260869565217391
|
0.0319148936170213
|
|
GO:CC
|
GO:1990111
|
spermatoproteasome complex
|
4.17900345466729e-07
|
5
|
40
|
3
|
0.075
|
0.6
|
|
GO:CC
|
GO:0012505
|
endomembrane system
|
1.03024321658712e-06
|
4640
|
28
|
20
|
0.714285714285714
|
0.00431034482758621
|
|
GO:CC
|
GO:0000502
|
proteasome complex
|
1.22676085531479e-06
|
66
|
40
|
5
|
0.125
|
0.0757575757575758
|
|
GO:CC
|
GO:1905369
|
endopeptidase complex
|
1.87535853812794e-06
|
72
|
40
|
5
|
0.125
|
0.0694444444444444
|
|
GO:CC
|
GO:0019774
|
proteasome core complex, beta-subunit complex
|
6.35368967759437e-06
|
11
|
40
|
3
|
0.075
|
0.272727272727273
|
|
GO:CC
|
GO:1905368
|
peptidase complex
|
7.56652256050001e-06
|
96
|
40
|
5
|
0.125
|
0.0520833333333333
|
|
GO:CC
|
GO:0043231
|
intracellular membrane-bounded organelle
|
1.02705893419704e-05
|
11296
|
25
|
25
|
1
|
0.00221317280453258
|
|
GO:CC
|
GO:0005576
|
extracellular region
|
1.13466164591805e-05
|
4567
|
123
|
53
|
0.430894308943089
|
0.0116049923363258
|
|
GO:CC
|
GO:0005769
|
early endosome
|
1.91299845253121e-05
|
378
|
23
|
6
|
0.260869565217391
|
0.0158730158730159
|
|
GO:CC
|
GO:0005737
|
cytoplasm
|
3.6695465306106e-05
|
11911
|
25
|
25
|
1
|
0.00209890017630761
|
|
GO:CC
|
GO:0016021
|
integral component of membrane
|
3.70360811980689e-05
|
5711
|
15
|
13
|
0.866666666666667
|
0.00227630887760462
|
|
GO:CC
|
GO:0031224
|
intrinsic component of membrane
|
5.03043257028569e-05
|
5868
|
15
|
13
|
0.866666666666667
|
0.00221540558963872
|
|
GO:CC
|
GO:0005886
|
plasma membrane
|
5.79159193073858e-05
|
5681
|
12
|
11
|
0.916666666666667
|
0.00193627882415068
|
|
GO:CC
|
GO:0005839
|
proteasome core complex
|
5.79159193073858e-05
|
23
|
40
|
3
|
0.075
|
0.130434782608696
|
|
GO:CC
|
GO:0043229
|
intracellular organelle
|
0.00011054956140915
|
12491
|
25
|
25
|
1
|
0.00200144103754703
|
|
GO:CC
|
GO:0071944
|
cell periphery
|
0.000113851266106373
|
6169
|
62
|
36
|
0.580645161290323
|
0.00583562976171178
|
|
GO:CC
|
GO:0043227
|
membrane-bounded organelle
|
0.000220303036671396
|
12858
|
25
|
25
|
1
|
0.00194431482345621
|
|
GO:CC
|
GO:0140534
|
endoplasmic reticulum protein-containing complex
|
0.000352818250073332
|
128
|
36
|
4
|
0.111111111111111
|
0.03125
|
|
GO:CC
|
GO:0031904
|
endosome lumen
|
0.000468266708239801
|
35
|
54
|
3
|
0.0555555555555556
|
0.0857142857142857
|
|
GO:CC
|
GO:0005887
|
integral component of plasma membrane
|
0.000756442316654179
|
1645
|
8
|
5
|
0.625
|
0.00303951367781155
|
|
GO:CC
|
GO:0072562
|
blood microparticle
|
0.00091212518053151
|
140
|
120
|
6
|
0.05
|
0.0428571428571429
|
|
GO:CC
|
GO:0031226
|
intrinsic component of plasma membrane
|
0.000930998915508441
|
1726
|
8
|
5
|
0.625
|
0.00289687137891078
|
|
GO:CC
|
GO:0043226
|
organelle
|
0.00170282170002271
|
14008
|
25
|
25
|
1
|
0.0017846944603084
|
|
GO:CC
|
GO:0140535
|
intracellular protein-containing complex
|
0.00411225266041388
|
767
|
21
|
5
|
0.238095238095238
|
0.00651890482398957
|
|
GO:CC
|
GO:0005622
|
intracellular anatomical structure
|
0.00636405270709423
|
14790
|
25
|
25
|
1
|
0.00169033130493577
|
|
GO:CC
|
GO:0043202
|
lysosomal lumen
|
0.00822892902672375
|
97
|
54
|
3
|
0.0555555555555556
|
0.0309278350515464
|
|
GO:CC
|
GO:0090734
|
site of DNA damage
|
0.00980947217672415
|
90
|
128
|
4
|
0.03125
|
0.0444444444444444
|
|
GO:CC
|
GO:0030667
|
secretory granule membrane
|
0.00980947217672415
|
303
|
19
|
3
|
0.157894736842105
|
0.0099009900990099
|
|
GO:CC
|
GO:0030141
|
secretory granule
|
0.0126556476912906
|
851
|
8
|
3
|
0.375
|
0.00352526439482961
|
|
GO:CC
|
GO:0016020
|
membrane
|
0.0135709209818275
|
9838
|
12
|
11
|
0.916666666666667
|
0.00111811343769059
|
|
GO:CC
|
GO:0099503
|
secretory vesicle
|
0.0200341060053747
|
1020
|
8
|
3
|
0.375
|
0.00294117647058824
|
|
GO:CC
|
GO:1902494
|
catalytic complex
|
0.045962160993234
|
1441
|
21
|
5
|
0.238095238095238
|
0.00346981263011797
|
|
GO:MF
|
GO:0042605
|
peptide antigen binding
|
1.34100603959402e-23
|
30
|
36
|
12
|
0.333333333333333
|
0.4
|
|
GO:MF
|
GO:0003823
|
antigen binding
|
4.65723898739062e-15
|
163
|
23
|
11
|
0.478260869565217
|
0.0674846625766871
|
|
GO:MF
|
GO:0042277
|
peptide binding
|
7.49283385503335e-14
|
314
|
23
|
12
|
0.521739130434783
|
0.0382165605095541
|
|
GO:MF
|
GO:0046977
|
TAP binding
|
3.27885814590976e-13
|
9
|
36
|
6
|
0.166666666666667
|
0.666666666666667
|
|
GO:MF
|
GO:0033218
|
amide binding
|
5.87524843979084e-13
|
389
|
23
|
12
|
0.521739130434783
|
0.0308483290488432
|
|
GO:MF
|
GO:0023023
|
MHC protein complex binding
|
1.69079204145285e-11
|
25
|
90
|
8
|
0.0888888888888889
|
0.32
|
|
GO:MF
|
GO:0023026
|
MHC class II protein complex binding
|
6.02505746723949e-11
|
17
|
90
|
7
|
0.0777777777777778
|
0.411764705882353
|
|
GO:MF
|
GO:0032395
|
MHC class II receptor activity
|
7.61851962150766e-09
|
8
|
90
|
5
|
0.0555555555555556
|
0.625
|
|
GO:MF
|
GO:0042608
|
T cell receptor binding
|
3.01979730638426e-08
|
11
|
23
|
4
|
0.173913043478261
|
0.363636363636364
|
|
GO:MF
|
GO:0048248
|
CXCR3 chemokine receptor binding
|
9.89688875056939e-08
|
5
|
86
|
4
|
0.0465116279069767
|
0.8
|
|
GO:MF
|
GO:0140375
|
immune receptor activity
|
1.05932677451108e-07
|
133
|
4
|
4
|
1
|
0.0300751879699248
|
|
GO:MF
|
GO:0042379
|
chemokine receptor binding
|
1.93857168070321e-06
|
72
|
135
|
8
|
0.0592592592592593
|
0.111111111111111
|
|
GO:MF
|
GO:0030881
|
beta-2-microglobulin binding
|
1.99145562331872e-06
|
7
|
23
|
3
|
0.130434782608696
|
0.428571428571429
|
|
GO:MF
|
GO:0008009
|
chemokine activity
|
1.99145562331872e-06
|
49
|
135
|
7
|
0.0518518518518519
|
0.142857142857143
|
|
GO:MF
|
GO:0046978
|
TAP1 binding
|
4.14053772380132e-06
|
6
|
36
|
3
|
0.0833333333333333
|
0.5
|
|
GO:MF
|
GO:0045236
|
CXCR chemokine receptor binding
|
4.31178189195261e-06
|
18
|
135
|
5
|
0.037037037037037
|
0.277777777777778
|
|
GO:MF
|
GO:0005102
|
signaling receptor binding
|
2.04313710396436e-05
|
1672
|
135
|
31
|
0.22962962962963
|
0.0185406698564593
|
|
GO:MF
|
GO:0042287
|
MHC protein binding
|
2.27343995852717e-05
|
39
|
36
|
4
|
0.111111111111111
|
0.102564102564103
|
|
GO:MF
|
GO:1990404
|
protein ADP-ribosylase activity
|
4.59487034760726e-05
|
16
|
111
|
4
|
0.036036036036036
|
0.25
|
|
GO:MF
|
GO:0004298
|
threonine-type endopeptidase activity
|
7.02232086974595e-05
|
14
|
40
|
3
|
0.075
|
0.214285714285714
|
|
GO:MF
|
GO:0042802
|
identical protein binding
|
8.93174932004972e-05
|
2067
|
137
|
34
|
0.248175182481752
|
0.0164489598451863
|
|
GO:MF
|
GO:0005126
|
cytokine receptor binding
|
0.000101765577024736
|
275
|
135
|
11
|
0.0814814814814815
|
0.04
|
|
GO:MF
|
GO:0042288
|
MHC class I protein binding
|
0.000133656112764514
|
20
|
36
|
3
|
0.0833333333333333
|
0.15
|
|
GO:MF
|
GO:0031730
|
CCR5 chemokine receptor binding
|
0.00026753809804291
|
8
|
123
|
3
|
0.024390243902439
|
0.375
|
|
GO:MF
|
GO:0070003
|
threonine-type peptidase activity
|
0.000302274330832288
|
24
|
40
|
3
|
0.075
|
0.125
|
|
GO:MF
|
GO:0003950
|
NAD+ ADP-ribosyltransferase activity
|
0.000305073828960446
|
27
|
111
|
4
|
0.036036036036036
|
0.148148148148148
|
|
GO:MF
|
GO:0042803
|
protein homodimerization activity
|
0.000314357359463691
|
676
|
131
|
16
|
0.122137404580153
|
0.0236686390532544
|
|
GO:MF
|
GO:0016763
|
transferase activity, transferring pentosyl groups
|
0.000314357359463691
|
56
|
111
|
5
|
0.045045045045045
|
0.0892857142857143
|
|
GO:MF
|
GO:0004888
|
transmembrane signaling receptor activity
|
0.000314357359463691
|
1269
|
4
|
4
|
1
|
0.00315208825847124
|
|
GO:MF
|
GO:0048020
|
CCR chemokine receptor binding
|
0.000351081348512658
|
48
|
135
|
5
|
0.037037037037037
|
0.104166666666667
|
|
GO:MF
|
GO:0004175
|
endopeptidase activity
|
0.000355834863555668
|
450
|
136
|
13
|
0.0955882352941176
|
0.0288888888888889
|
|
GO:MF
|
GO:0060089
|
molecular transducer activity
|
0.000461572856645642
|
1458
|
4
|
4
|
1
|
0.00274348422496571
|
|
GO:MF
|
GO:0038023
|
signaling receptor activity
|
0.000461572856645642
|
1458
|
4
|
4
|
1
|
0.00274348422496571
|
|
GO:MF
|
GO:0005125
|
cytokine activity
|
0.000623450940956867
|
235
|
135
|
9
|
0.0666666666666667
|
0.0382978723404255
|
|
GO:MF
|
GO:0008233
|
peptidase activity
|
0.000820329100585741
|
646
|
136
|
15
|
0.110294117647059
|
0.0232198142414861
|
|
GO:MF
|
GO:0044877
|
protein-containing complex binding
|
0.00192739177593524
|
1287
|
45
|
11
|
0.244444444444444
|
0.00854700854700855
|
|
GO:MF
|
GO:0048018
|
receptor ligand activity
|
0.00257585093526741
|
494
|
135
|
12
|
0.0888888888888889
|
0.0242914979757085
|
|
GO:MF
|
GO:0004252
|
serine-type endopeptidase activity
|
0.00260827735667843
|
171
|
136
|
7
|
0.0514705882352941
|
0.0409356725146199
|
|
GO:MF
|
GO:0001664
|
G protein-coupled receptor binding
|
0.00267527378249877
|
292
|
135
|
9
|
0.0666666666666667
|
0.0308219178082192
|
|
GO:MF
|
GO:0030546
|
signaling receptor activator activity
|
0.0026932665318106
|
501
|
135
|
12
|
0.0888888888888889
|
0.0239520958083832
|
|
GO:MF
|
GO:0008236
|
serine-type peptidase activity
|
0.00431518609441441
|
189
|
136
|
7
|
0.0514705882352941
|
0.037037037037037
|
|
GO:MF
|
GO:0017171
|
serine hydrolase activity
|
0.00479735719008619
|
193
|
136
|
7
|
0.0514705882352941
|
0.0362694300518135
|
|
GO:MF
|
GO:0098772
|
molecular function regulator
|
0.00502360966463116
|
2363
|
140
|
32
|
0.228571428571429
|
0.0135421074904782
|
|
GO:MF
|
GO:0030545
|
receptor regulator activity
|
0.00511136800481163
|
545
|
135
|
12
|
0.0888888888888889
|
0.0220183486238532
|
|
GO:MF
|
GO:0005164
|
tumor necrosis factor receptor binding
|
0.00611156339123549
|
32
|
105
|
3
|
0.0285714285714286
|
0.09375
|
|
GO:MF
|
GO:0140096
|
catalytic activity, acting on a protein
|
0.00665549327838424
|
2332
|
139
|
31
|
0.223021582733813
|
0.0132933104631218
|
|
GO:MF
|
GO:0061135
|
endopeptidase regulator activity
|
0.0086518024351163
|
188
|
21
|
3
|
0.142857142857143
|
0.0159574468085106
|
|
GO:MF
|
GO:0005525
|
GTP binding
|
0.00898173727536685
|
381
|
80
|
7
|
0.0875
|
0.0183727034120735
|
|
GO:MF
|
GO:0032550
|
purine ribonucleoside binding
|
0.00939183207244835
|
386
|
80
|
7
|
0.0875
|
0.0181347150259067
|
|
GO:MF
|
GO:0046983
|
protein dimerization activity
|
0.00939183207244835
|
1069
|
131
|
17
|
0.129770992366412
|
0.0159027128157156
|
|
GO:MF
|
GO:0032549
|
ribonucleoside binding
|
0.00953361786807994
|
389
|
80
|
7
|
0.0875
|
0.0179948586118252
|
|
GO:MF
|
GO:0001883
|
purine nucleoside binding
|
0.00953361786807994
|
389
|
80
|
7
|
0.0875
|
0.0179948586118252
|
|
GO:MF
|
GO:0001882
|
nucleoside binding
|
0.0102571122908542
|
395
|
80
|
7
|
0.0875
|
0.0177215189873418
|
|
GO:MF
|
GO:0043394
|
proteoglycan binding
|
0.010564226549904
|
36
|
120
|
3
|
0.025
|
0.0833333333333333
|
|
GO:MF
|
GO:0032561
|
guanyl ribonucleotide binding
|
0.0110252004958694
|
403
|
80
|
7
|
0.0875
|
0.0173697270471464
|
|
GO:MF
|
GO:0019001
|
guanyl nucleotide binding
|
0.0110252004958694
|
403
|
80
|
7
|
0.0875
|
0.0173697270471464
|
|
GO:MF
|
GO:0061134
|
peptidase regulator activity
|
0.01188222070717
|
224
|
21
|
3
|
0.142857142857143
|
0.0133928571428571
|
|
GO:MF
|
GO:0042056
|
chemoattractant activity
|
0.0143879550489605
|
37
|
135
|
3
|
0.0222222222222222
|
0.0810810810810811
|
|
GO:MF
|
GO:0032813
|
tumor necrosis factor receptor superfamily binding
|
0.0154304631352756
|
49
|
105
|
3
|
0.0285714285714286
|
0.0612244897959184
|
|
GO:MF
|
GO:0016787
|
hydrolase activity
|
0.0168753259674024
|
2615
|
129
|
30
|
0.232558139534884
|
0.011472275334608
|
|
GO:MF
|
GO:0005515
|
protein binding
|
0.0212989906993251
|
14767
|
71
|
65
|
0.915492957746479
|
0.00440170650775378
|
|
GO:MF
|
GO:0097367
|
carbohydrate derivative binding
|
0.021631705265114
|
2285
|
88
|
20
|
0.227272727272727
|
0.0087527352297593
|
|
GO:MF
|
GO:0005539
|
glycosaminoglycan binding
|
0.0223982369485956
|
238
|
120
|
6
|
0.05
|
0.0252100840336134
|
|
GO:MF
|
GO:0008201
|
heparin binding
|
0.0227447084760032
|
168
|
120
|
5
|
0.0416666666666667
|
0.0297619047619048
|
|
GO:MF
|
GO:0019955
|
cytokine binding
|
0.0255376306253019
|
138
|
97
|
4
|
0.0412371134020619
|
0.0289855072463768
|
|
GO:MF
|
GO:0003924
|
GTPase activity
|
0.0418370417458506
|
328
|
106
|
6
|
0.0566037735849057
|
0.0182926829268293
|
|
GO:MF
|
GO:0003714
|
transcription corepressor activity
|
0.0461917448098832
|
187
|
89
|
4
|
0.0449438202247191
|
0.0213903743315508
|
|
HP
|
HP:0011122
|
Abnormality of skin physiology
|
2.31717580895823e-05
|
430
|
13
|
10
|
0.769230769230769
|
0.0232558139534884
|
|
HP
|
HP:0011123
|
Inflammatory abnormality of the skin
|
6.78537850994437e-05
|
367
|
13
|
9
|
0.692307692307692
|
0.0245231607629428
|
|
HP
|
HP:0003565
|
Elevated erythrocyte sedimentation rate
|
6.78537850994437e-05
|
48
|
13
|
5
|
0.384615384615385
|
0.104166666666667
|
|
HP
|
HP:0025021
|
Abnormal erythrocyte sedimentation rate
|
6.78537850994437e-05
|
49
|
13
|
5
|
0.384615384615385
|
0.102040816326531
|
|
HP
|
HP:0100533
|
Inflammatory abnormality of the eye
|
8.60918544132623e-05
|
193
|
13
|
7
|
0.538461538461538
|
0.0362694300518135
|
|
HP
|
HP:0003765
|
Psoriasiform dermatitis
|
8.60918544132623e-05
|
25
|
11
|
4
|
0.363636363636364
|
0.16
|
|
HP
|
HP:0001701
|
Pericarditis
|
8.60918544132623e-05
|
34
|
9
|
4
|
0.444444444444444
|
0.117647058823529
|
|
HP
|
HP:0045073
|
Serositis
|
0.000131246186708986
|
40
|
9
|
4
|
0.444444444444444
|
0.1
|
|
HP
|
HP:0033331
|
Acute phase response
|
0.000131246186708986
|
65
|
13
|
5
|
0.384615384615385
|
0.0769230769230769
|
|
HP
|
HP:0000988
|
Skin rash
|
0.000201293752413128
|
114
|
9
|
5
|
0.555555555555556
|
0.043859649122807
|
|
HP
|
HP:0100749
|
Chest pain
|
0.000233523002328303
|
93
|
11
|
5
|
0.454545454545455
|
0.0537634408602151
|
|
HP
|
HP:0002113
|
Pulmonary infiltrates
|
0.00031173696776463
|
53
|
9
|
4
|
0.444444444444444
|
0.0754716981132075
|
|
HP
|
HP:0002829
|
Arthralgia
|
0.00037211225353837
|
161
|
13
|
6
|
0.461538461538462
|
0.0372670807453416
|
|
HP
|
HP:0002105
|
Hemoptysis
|
0.000413068523823357
|
59
|
9
|
4
|
0.444444444444444
|
0.0677966101694915
|
|
HP
|
HP:0000509
|
Conjunctivitis
|
0.000598650859485448
|
98
|
13
|
5
|
0.384615384615385
|
0.0510204081632653
|
|
HP
|
HP:0001697
|
Abnormal pericardium morphology
|
0.000603783226881345
|
68
|
9
|
4
|
0.444444444444444
|
0.0588235294117647
|
|
HP
|
HP:0025337
|
Red eye
|
0.000603783226881345
|
100
|
13
|
5
|
0.384615384615385
|
0.05
|
|
HP
|
HP:0012735
|
Cough
|
0.000645998838690602
|
128
|
11
|
5
|
0.454545454545455
|
0.0390625
|
|
HP
|
HP:0031983
|
Abnormal pulmonary thoracic imaging finding
|
0.000645998838690602
|
71
|
9
|
4
|
0.444444444444444
|
0.0563380281690141
|
|
HP
|
HP:0001287
|
Meningitis
|
0.000645998838690602
|
72
|
9
|
4
|
0.444444444444444
|
0.0555555555555556
|
|
HP
|
HP:0011227
|
Elevated C-reactive protein level
|
0.000835632863842384
|
51
|
13
|
4
|
0.307692307692308
|
0.0784313725490196
|
|
HP
|
HP:0002091
|
Restrictive ventilatory defect
|
0.000857363763636463
|
80
|
9
|
4
|
0.444444444444444
|
0.05
|
|
HP
|
HP:0032436
|
Abnormal C-reactive protein level
|
0.000857363763636463
|
52
|
13
|
4
|
0.307692307692308
|
0.0769230769230769
|
|
HP
|
HP:0002102
|
Pleuritis
|
0.000935494672340329
|
25
|
9
|
3
|
0.333333333333333
|
0.12
|
|
HP
|
HP:0002103
|
Abnormal pleura morphology
|
0.000958823769091669
|
84
|
9
|
4
|
0.444444444444444
|
0.0476190476190476
|
|
HP
|
HP:0011450
|
Unusual CNS infection
|
0.00106069006659212
|
87
|
9
|
4
|
0.444444444444444
|
0.0459770114942529
|
|
HP
|
HP:0001824
|
Weight loss
|
0.0010921415979816
|
273
|
11
|
6
|
0.545454545454545
|
0.021978021978022
|
|
HP
|
HP:0200042
|
Skin ulcer
|
0.0010921415979816
|
115
|
14
|
5
|
0.357142857142857
|
0.0434782608695652
|
|
HP
|
HP:0032158
|
Unusual infection by anatomical site
|
0.00113771864525245
|
91
|
9
|
4
|
0.444444444444444
|
0.043956043956044
|
|
HP
|
HP:0000246
|
Sinusitis
|
0.00150941827811339
|
125
|
14
|
5
|
0.357142857142857
|
0.04
|
|
HP
|
HP:0000502
|
Abnormal conjunctiva morphology
|
0.00172776385366944
|
141
|
13
|
5
|
0.384615384615385
|
0.0354609929078014
|
|
HP
|
HP:0001097
|
Keratoconjunctivitis sicca
|
0.00174300875066392
|
39
|
8
|
3
|
0.375
|
0.0769230769230769
|
|
HP
|
HP:0032016
|
Abnormal sputum
|
0.00174300875066392
|
106
|
9
|
4
|
0.444444444444444
|
0.0377358490566038
|
|
HP
|
HP:0001426
|
Multifactorial inheritance
|
0.00174300875066392
|
84
|
11
|
4
|
0.363636363636364
|
0.0476190476190476
|
|
HP
|
HP:0000763
|
Sensory neuropathy
|
0.00195310554557297
|
182
|
11
|
5
|
0.454545454545455
|
0.0274725274725275
|
|
HP
|
HP:0001096
|
Keratoconjunctivitis
|
0.00214595162194157
|
43
|
8
|
3
|
0.375
|
0.0697674418604651
|
|
HP
|
HP:0000245
|
Abnormal paranasal sinus morphology
|
0.00222916513084722
|
143
|
14
|
5
|
0.357142857142857
|
0.034965034965035
|
|
HP
|
HP:0012647
|
Abnormal inflammatory response
|
0.00229209870703601
|
914
|
14
|
10
|
0.714285714285714
|
0.0109409190371991
|
|
HP
|
HP:0012649
|
Increased inflammatory response
|
0.00229209870703601
|
914
|
14
|
10
|
0.714285714285714
|
0.0109409190371991
|
|
HP
|
HP:0100820
|
Glomerulopathy
|
0.00262290654862846
|
42
|
9
|
3
|
0.333333333333333
|
0.0714285714285714
|
|
HP
|
HP:0030878
|
Abnormality on pulmonary function testing
|
0.00296168186484014
|
128
|
9
|
4
|
0.444444444444444
|
0.03125
|
|
HP
|
HP:0002719
|
Recurrent infections
|
0.00321962653386963
|
745
|
14
|
9
|
0.642857142857143
|
0.0120805369127517
|
|
HP
|
HP:0001945
|
Fever
|
0.00331116895854205
|
294
|
13
|
6
|
0.461538461538462
|
0.0204081632653061
|
|
HP
|
HP:0000969
|
Edema
|
0.00372261005080971
|
489
|
9
|
6
|
0.666666666666667
|
0.0122699386503067
|
|
HP
|
HP:0002239
|
Gastrointestinal hemorrhage
|
0.00372261005080971
|
141
|
9
|
4
|
0.444444444444444
|
0.0283687943262411
|
|
HP
|
HP:0100758
|
Gangrene
|
0.0041165419167552
|
52
|
9
|
3
|
0.333333333333333
|
0.0576923076923077
|
|
HP
|
HP:0032101
|
Unusual infection
|
0.00468753962060952
|
792
|
14
|
9
|
0.642857142857143
|
0.0113636363636364
|
|
HP
|
HP:0000822
|
Hypertension
|
0.00482775648927618
|
306
|
9
|
5
|
0.555555555555556
|
0.0163398692810458
|
|
HP
|
HP:0000873
|
Diabetes insipidus
|
0.0051215250880661
|
57
|
9
|
3
|
0.333333333333333
|
0.0526315789473684
|
|
HP
|
HP:0100539
|
Periorbital edema
|
0.0057583110319992
|
60
|
9
|
3
|
0.333333333333333
|
0.05
|
|
HP
|
HP:0000491
|
Keratitis
|
0.00660542013269737
|
74
|
8
|
3
|
0.375
|
0.0405405405405405
|
|
HP
|
HP:0011032
|
Abnormality of fluid regulation
|
0.00660542013269737
|
562
|
9
|
6
|
0.666666666666667
|
0.0106761565836299
|
|
HP
|
HP:0010783
|
Erythema
|
0.00660542013269737
|
113
|
13
|
4
|
0.307692307692308
|
0.0353982300884956
|
|
HP
|
HP:0001369
|
Arthritis
|
0.00660542013269737
|
196
|
8
|
4
|
0.5
|
0.0204081632653061
|
|
HP
|
HP:0002633
|
Vasculitis
|
0.00660542013269737
|
65
|
9
|
3
|
0.333333333333333
|
0.0461538461538462
|
|
HP
|
HP:0032263
|
Increased blood pressure
|
0.00752700018384323
|
350
|
9
|
5
|
0.555555555555556
|
0.0142857142857143
|
|
HP
|
HP:0002206
|
Pulmonary fibrosis
|
0.00752700018384323
|
69
|
9
|
3
|
0.333333333333333
|
0.0434782608695652
|
|
HP
|
HP:0000282
|
Facial edema
|
0.00752700018384323
|
69
|
9
|
3
|
0.333333333333333
|
0.0434782608695652
|
|
HP
|
HP:0030355
|
Abnormal serum interferon-gamma level
|
0.00755389277515176
|
11
|
56
|
3
|
0.0535714285714286
|
0.272727272727273
|
|
HP
|
HP:0030354
|
Abnormal serum interferon level
|
0.00755389277515176
|
11
|
56
|
3
|
0.0535714285714286
|
0.272727272727273
|
|
HP
|
HP:0002960
|
Autoimmunity
|
0.0090968125309544
|
170
|
73
|
10
|
0.136986301369863
|
0.0588235294117647
|
|
HP
|
HP:0001733
|
Pancreatitis
|
0.0090968125309544
|
76
|
9
|
3
|
0.333333333333333
|
0.0394736842105263
|
|
HP
|
HP:0200034
|
Papule
|
0.00964328965589076
|
78
|
9
|
3
|
0.333333333333333
|
0.0384615384615385
|
|
HP
|
HP:0004936
|
Venous thrombosis
|
0.0107390565599849
|
82
|
9
|
3
|
0.333333333333333
|
0.0365853658536585
|
|
HP
|
HP:0012575
|
Abnormal nephron morphology
|
0.0107772303330332
|
208
|
9
|
4
|
0.444444444444444
|
0.0192307692307692
|
|
HP
|
HP:0004370
|
Abnormality of temperature regulation
|
0.0107772303330332
|
398
|
13
|
6
|
0.461538461538462
|
0.0150753768844221
|
|
HP
|
HP:0000022
|
Abnormality of male internal genitalia
|
0.0114960795121083
|
57
|
13
|
3
|
0.230769230769231
|
0.0526315789473684
|
|
HP
|
HP:0011495
|
Abnormal corneal epithelium morphology
|
0.0135210758456252
|
103
|
8
|
3
|
0.375
|
0.029126213592233
|
|
HP
|
HP:0030972
|
Abnormal systemic blood pressure
|
0.0145885063216851
|
423
|
9
|
5
|
0.555555555555556
|
0.0118203309692671
|
|
HP
|
HP:0011947
|
Respiratory tract infection
|
0.0148704818268207
|
558
|
14
|
7
|
0.5
|
0.0125448028673835
|
|
HP
|
HP:0008066
|
Abnormal blistering of the skin
|
0.0160532106565639
|
78
|
11
|
3
|
0.272727272727273
|
0.0384615384615385
|
|
HP
|
HP:0002637
|
Cerebral ischemia
|
0.0165212950728408
|
99
|
9
|
3
|
0.333333333333333
|
0.0303030303030303
|
|
HP
|
HP:0033401
|
Tissue ischemia
|
0.0165212950728408
|
99
|
9
|
3
|
0.333333333333333
|
0.0303030303030303
|
|
HP
|
HP:0012531
|
Pain
|
0.0179955065851962
|
636
|
13
|
7
|
0.538461538461538
|
0.0110062893081761
|
|
HP
|
HP:0030163
|
Abnormal vascular physiology
|
0.0186933960722413
|
248
|
9
|
4
|
0.444444444444444
|
0.0161290322580645
|
|
HP
|
HP:0002027
|
Abdominal pain
|
0.0192381437894662
|
251
|
9
|
4
|
0.444444444444444
|
0.0159362549800797
|
|
HP
|
HP:0004374
|
Hemiplegia/hemiparesis
|
0.0192381437894662
|
199
|
11
|
4
|
0.363636363636364
|
0.0201005025125628
|
|
HP
|
HP:0000488
|
Retinopathy
|
0.0199190567077615
|
108
|
9
|
3
|
0.333333333333333
|
0.0277777777777778
|
|
HP
|
HP:0009830
|
Peripheral neuropathy
|
0.0233267291320862
|
588
|
11
|
6
|
0.545454545454545
|
0.0102040816326531
|
|
HP
|
HP:0010876
|
Abnormal circulating protein concentration
|
0.0233267291320862
|
480
|
13
|
6
|
0.461538461538462
|
0.0125
|
|
HP
|
HP:0003401
|
Paresthesia
|
0.0235231040756702
|
96
|
11
|
3
|
0.272727272727273
|
0.03125
|
|
HP
|
HP:0008047
|
Abnormality of the vasculature of the eye
|
0.0235231040756702
|
319
|
8
|
4
|
0.5
|
0.0125391849529781
|
|
HP
|
HP:0001977
|
Abnormal thrombosis
|
0.0235231040756702
|
120
|
9
|
3
|
0.333333333333333
|
0.025
|
|
HP
|
HP:0011029
|
Internal hemorrhage
|
0.0245196611156925
|
282
|
9
|
4
|
0.444444444444444
|
0.0141843971631206
|
|
HP
|
HP:0011276
|
Vascular skin abnormality
|
0.0245196611156925
|
493
|
13
|
6
|
0.461538461538462
|
0.0121703853955375
|
|
HP
|
HP:0031910
|
Abnormal cranial nerve physiology
|
0.0249359594477871
|
288
|
9
|
4
|
0.444444444444444
|
0.0138888888888889
|
|
HP
|
HP:0001881
|
Abnormal leukocyte morphology
|
0.0249359594477871
|
510
|
9
|
5
|
0.555555555555556
|
0.00980392156862745
|
|
HP
|
HP:0002716
|
Lymphadenopathy
|
0.0249359594477871
|
187
|
13
|
4
|
0.307692307692308
|
0.0213903743315508
|
|
HP
|
HP:0010987
|
Abnormal cellular immune system morphology
|
0.0249359594477871
|
510
|
9
|
5
|
0.555555555555556
|
0.00980392156862745
|
|
HP
|
HP:0006824
|
Cranial nerve paralysis
|
0.0249359594477871
|
287
|
9
|
4
|
0.444444444444444
|
0.0139372822299652
|
|
HP
|
HP:0010978
|
Abnormality of immune system physiology
|
0.0249626347698135
|
1346
|
14
|
10
|
0.714285714285714
|
0.00742942050520059
|
|
HP
|
HP:0025142
|
Constitutional symptom
|
0.0253357443683905
|
939
|
13
|
8
|
0.615384615384615
|
0.00851970181043664
|
|
HP
|
HP:0025323
|
Abnormal arterial physiology
|
0.0258289563698038
|
130
|
9
|
3
|
0.333333333333333
|
0.0230769230769231
|
|
HP
|
HP:0002733
|
Abnormality of the lymph nodes
|
0.0258289563698038
|
196
|
13
|
4
|
0.307692307692308
|
0.0204081632653061
|
|
HP
|
HP:0002205
|
Recurrent respiratory infections
|
0.0258289563698038
|
468
|
14
|
6
|
0.428571428571429
|
0.0128205128205128
|
|
HP
|
HP:0012733
|
Macule
|
0.0258289563698038
|
106
|
11
|
3
|
0.272727272727273
|
0.0283018867924528
|
|
HP
|
HP:0001744
|
Splenomegaly
|
0.0259690671659964
|
345
|
8
|
4
|
0.5
|
0.0115942028985507
|
|
HP
|
HP:0032251
|
Abnormal immune system morphology
|
0.0262615987427475
|
532
|
9
|
5
|
0.555555555555556
|
0.0093984962406015
|
|
HP
|
HP:0012091
|
Abnormality of pancreas physiology
|
0.0267699507401918
|
136
|
9
|
3
|
0.333333333333333
|
0.0220588235294118
|
|
HP
|
HP:0010701
|
Abnormal immunoglobulin level
|
0.0268260794361516
|
219
|
6
|
3
|
0.5
|
0.0136986301369863
|
|
HP
|
HP:0010702
|
Increased circulating antibody level
|
0.0268260794361516
|
92
|
13
|
3
|
0.230769230769231
|
0.0326086956521739
|
|
HP
|
HP:0011028
|
Abnormality of blood circulation
|
0.0268260794361516
|
306
|
9
|
4
|
0.444444444444444
|
0.0130718954248366
|
|
HP
|
HP:0005372
|
Abnormality of B cell physiology
|
0.0269858913170111
|
220
|
6
|
3
|
0.5
|
0.0136363636363636
|
|
HP
|
HP:0000790
|
Hematuria
|
0.0272883993241668
|
140
|
9
|
3
|
0.333333333333333
|
0.0214285714285714
|
|
HP
|
HP:0012614
|
Abnormal urine cytology
|
0.0272883993241668
|
140
|
9
|
3
|
0.333333333333333
|
0.0214285714285714
|
|
HP
|
HP:0006530
|
Abnormal pulmonary Interstitial morphology
|
0.0272883993241668
|
140
|
9
|
3
|
0.333333333333333
|
0.0214285714285714
|
|
HP
|
HP:0010974
|
Abnormal myeloid leukocyte morphology
|
0.0272883993241668
|
312
|
9
|
4
|
0.444444444444444
|
0.0128205128205128
|
|
HP
|
HP:0003326
|
Myalgia
|
0.0272883993241668
|
140
|
9
|
3
|
0.333333333333333
|
0.0214285714285714
|
|
HP
|
HP:0012252
|
Abnormal respiratory system morphology
|
0.0290080149425047
|
1137
|
14
|
9
|
0.642857142857143
|
0.0079155672823219
|
|
HP
|
HP:0002110
|
Bronchiectasis
|
0.0290080149425047
|
119
|
36
|
5
|
0.138888888888889
|
0.0420168067226891
|
|
HP
|
HP:0100755
|
Abnormality of salivation
|
0.0320443530933165
|
172
|
8
|
3
|
0.375
|
0.0174418604651163
|
|
HP
|
HP:0011112
|
Abnormality of serum cytokine level
|
0.0323510647753324
|
23
|
56
|
3
|
0.0535714285714286
|
0.130434782608696
|
|
HP
|
HP:0002354
|
Memory impairment
|
0.0332102242365564
|
123
|
11
|
3
|
0.272727272727273
|
0.024390243902439
|
|
HP
|
HP:0012378
|
Fatigue
|
0.0332210509715629
|
335
|
9
|
4
|
0.444444444444444
|
0.0119402985074627
|
|
HP
|
HP:0031409
|
Abnormal lymphocyte physiology
|
0.0334044761543684
|
246
|
6
|
3
|
0.5
|
0.0121951219512195
|
|
HP
|
HP:0000083
|
Renal insufficiency
|
0.0335232094106596
|
337
|
9
|
4
|
0.444444444444444
|
0.0118694362017804
|
|
HP
|
HP:0005368
|
Abnormality of humoral immunity
|
0.034149186195781
|
249
|
6
|
3
|
0.5
|
0.0120481927710843
|
|
HP
|
HP:0000095
|
Abnormal renal glomerulus morphology
|
0.0345864346060774
|
158
|
9
|
3
|
0.333333333333333
|
0.0189873417721519
|
|
HP
|
HP:0011111
|
Abnormality of immune serum protein physiology
|
0.0345864346060774
|
24
|
56
|
3
|
0.0535714285714286
|
0.125
|
|
HP
|
HP:0031263
|
Abnormal renal corpuscle morphology
|
0.0345864346060774
|
158
|
9
|
3
|
0.333333333333333
|
0.0189873417721519
|
|
HP
|
HP:0011675
|
Arrhythmia
|
0.0346079789710122
|
381
|
13
|
5
|
0.384615384615385
|
0.0131233595800525
|
|
HP
|
HP:0000093
|
Proteinuria
|
0.0400738494765367
|
168
|
9
|
3
|
0.333333333333333
|
0.0178571428571429
|
|
HP
|
HP:0002088
|
Abnormal lung morphology
|
0.0400738494765367
|
960
|
14
|
8
|
0.571428571428571
|
0.00833333333333333
|
|
HP
|
HP:0012211
|
Abnormal renal physiology
|
0.0400738494765367
|
614
|
9
|
5
|
0.555555555555556
|
0.00814332247557003
|
|
HP
|
HP:0002024
|
Malabsorption
|
0.0414587723596119
|
195
|
8
|
3
|
0.375
|
0.0153846153846154
|
|
HP
|
HP:0002037
|
Inflammation of the large intestine
|
0.0434884538502253
|
62
|
24
|
3
|
0.125
|
0.0483870967741935
|
|
HP
|
HP:0011799
|
Abnormality of facial soft tissue
|
0.0445000973545186
|
413
|
13
|
5
|
0.384615384615385
|
0.0121065375302663
|
|
HP
|
HP:0020129
|
Abnormal urine protein level
|
0.0445000973545186
|
181
|
9
|
3
|
0.333333333333333
|
0.0165745856353591
|
|
HP
|
HP:0002087
|
Abnormality of the upper respiratory tract
|
0.0446143276651034
|
382
|
14
|
5
|
0.357142857142857
|
0.0130890052356021
|
|
HP
|
HP:0025426
|
Abnormal bronchus morphology
|
0.0454798529096364
|
140
|
36
|
5
|
0.138888888888889
|
0.0357142857142857
|
|
HP
|
HP:0030956
|
Abnormality of cardiovascular system electrophysiology
|
0.0468984631474403
|
424
|
13
|
5
|
0.384615384615385
|
0.0117924528301887
|
|
HP
|
HP:0011035
|
Abnormal renal cortex morphology
|
0.0468984631474403
|
187
|
9
|
3
|
0.333333333333333
|
0.0160427807486631
|
|
HP
|
HP:0000951
|
Abnormality of the skin
|
0.0472747816541248
|
1868
|
9
|
8
|
0.888888888888889
|
0.00428265524625268
|
|
HP
|
HP:0031815
|
Abnormal oral physiology
|
0.0484288482226132
|
218
|
8
|
3
|
0.375
|
0.0137614678899083
|
|
HP
|
HP:0000388
|
Otitis media
|
0.0499750468053343
|
245
|
14
|
4
|
0.285714285714286
|
0.0163265306122449
|
|
HPA
|
HPA:0310443
|
lymph node; non-germinal center cells[High]
|
5.02000652921445e-05
|
1169
|
84
|
27
|
0.321428571428571
|
0.0230966638152267
|
|
HPA
|
HPA:0530722
|
spleen; cells in white pulp[≥Medium]
|
0.00043117988413457
|
2645
|
26
|
18
|
0.692307692307692
|
0.00680529300567108
|
|
HPA
|
HPA:0310433
|
lymph node; germinal center cells[High]
|
0.000501976919357029
|
908
|
26
|
11
|
0.423076923076923
|
0.0121145374449339
|
|
HPA
|
HPA:0310432
|
lymph node; germinal center cells[≥Medium]
|
0.000715823380109997
|
3209
|
26
|
19
|
0.730769230769231
|
0.00592084761607978
|
|
HPA
|
HPA:0600443
|
tonsil; non-germinal center cells[High]
|
0.00103682288810088
|
1271
|
12
|
8
|
0.666666666666667
|
0.00629425649095201
|
|
HPA
|
HPA:0310442
|
lymph node; non-germinal center cells[≥Medium]
|
0.00110958917418223
|
3875
|
14
|
13
|
0.928571428571429
|
0.00335483870967742
|
|
HPA
|
HPA:0030072
|
appendix; lymphoid tissue[≥Medium]
|
0.00110958917418223
|
3131
|
14
|
12
|
0.857142857142857
|
0.00383264132864899
|
|
HPA
|
HPA:0600432
|
tonsil; germinal center cells[≥Medium]
|
0.00123941018626317
|
3988
|
14
|
13
|
0.928571428571429
|
0.00325977933801404
|
|
HPA
|
HPA:0600442
|
tonsil; non-germinal center cells[≥Medium]
|
0.00146015019018083
|
4079
|
14
|
13
|
0.928571428571429
|
0.00318705565089483
|
|
HPA
|
HPA:0600433
|
tonsil; germinal center cells[High]
|
0.00180578904220915
|
1179
|
11
|
7
|
0.636363636363636
|
0.00593723494486853
|
|
HPA
|
HPA:0530721
|
spleen; cells in white pulp[≥Low]
|
0.00198680585889477
|
4249
|
14
|
13
|
0.928571428571429
|
0.00305954342198164
|
|
HPA
|
HPA:0530712
|
spleen; cells in red pulp[≥Medium]
|
0.00337748386435472
|
3049
|
26
|
17
|
0.653846153846154
|
0.0055755985569039
|
|
HPA
|
HPA:0300412
|
lung; macrophages[≥Medium]
|
0.003736977670397
|
4732
|
29
|
23
|
0.793103448275862
|
0.00486052409129332
|
|
HPA
|
HPA:0300413
|
lung; macrophages[High]
|
0.00441690842280475
|
1224
|
20
|
9
|
0.45
|
0.00735294117647059
|
|
HPA
|
HPA:0211092
|
fallopian tube; non-ciliated cells[≥Medium]
|
0.00441690842280475
|
328
|
10
|
4
|
0.4
|
0.0121951219512195
|
|
HPA
|
HPA:0310431
|
lymph node; germinal center cells[≥Low]
|
0.00700708801956762
|
4935
|
26
|
21
|
0.807692307692308
|
0.00425531914893617
|
|
HPA
|
HPA:0030071
|
appendix; lymphoid tissue[≥Low]
|
0.00809713021808194
|
4976
|
14
|
13
|
0.928571428571429
|
0.00261254019292604
|
|
HPA
|
HPA:0211091
|
fallopian tube; non-ciliated cells[≥Low]
|
0.0113391848284426
|
456
|
10
|
4
|
0.4
|
0.0087719298245614
|
|
HPA
|
HPA:0530711
|
spleen; cells in red pulp[≥Low]
|
0.0118433244638701
|
5195
|
14
|
13
|
0.928571428571429
|
0.00250240615976901
|
|
HPA
|
HPA:0530723
|
spleen; cells in white pulp[High]
|
0.0138083873432713
|
811
|
11
|
5
|
0.454545454545455
|
0.0061652281134402
|
|
HPA
|
HPA:0460642
|
skin 1; Langerhans[≥Medium]
|
0.0198251801453468
|
3123
|
14
|
10
|
0.714285714285714
|
0.0032020493115594
|
|
HPA
|
HPA:0460643
|
skin 1; Langerhans[High]
|
0.0261612996142597
|
734
|
14
|
5
|
0.357142857142857
|
0.00681198910081744
|
|
HPA
|
HPA:0270352
|
kidney; cells in glomeruli[≥Medium]
|
0.0289604563072059
|
2914
|
20
|
12
|
0.6
|
0.00411805078929307
|
|
HPA
|
HPA:0530000
|
spleen
|
0.0291204565510125
|
5756
|
14
|
13
|
0.928571428571429
|
0.0022585128561501
|
|
HPA
|
HPA:0600431
|
tonsil; germinal center cells[≥Low]
|
0.0339188951518282
|
5845
|
14
|
13
|
0.928571428571429
|
0.00222412318220701
|
|
HPA
|
HPA:0310441
|
lymph node; non-germinal center cells[≥Low]
|
0.0379338716108611
|
5915
|
14
|
13
|
0.928571428571429
|
0.0021978021978022
|
|
HPA
|
HPA:0600441
|
tonsil; non-germinal center cells[≥Low]
|
0.0407995265654715
|
5966
|
14
|
13
|
0.928571428571429
|
0.00217901441501844
|
|
KEGG
|
KEGG:04612
|
Antigen processing and presentation
|
3.41457003466494e-31
|
69
|
25
|
18
|
0.72
|
0.260869565217391
|
|
KEGG
|
KEGG:05330
|
Allograft rejection
|
3.18406203468232e-24
|
34
|
25
|
13
|
0.52
|
0.382352941176471
|
|
KEGG
|
KEGG:05332
|
Graft-versus-host disease
|
8.1147967357042e-24
|
37
|
25
|
13
|
0.52
|
0.351351351351351
|
|
KEGG
|
KEGG:04940
|
Type I diabetes mellitus
|
2.04713392577899e-23
|
40
|
25
|
13
|
0.52
|
0.325
|
|
KEGG
|
KEGG:05320
|
Autoimmune thyroid disease
|
3.52914000901113e-22
|
49
|
25
|
13
|
0.52
|
0.26530612244898
|
|
KEGG
|
KEGG:05416
|
Viral myocarditis
|
2.0959362258473e-21
|
56
|
25
|
13
|
0.52
|
0.232142857142857
|
|
KEGG
|
KEGG:05169
|
Epstein-Barr virus infection
|
3.07914858897212e-19
|
198
|
25
|
16
|
0.64
|
0.0808080808080808
|
|
KEGG
|
KEGG:04145
|
Phagosome
|
1.48791190430316e-17
|
147
|
25
|
14
|
0.56
|
0.0952380952380952
|
|
KEGG
|
KEGG:05168
|
Herpes simplex virus 1 infection
|
5.35932252677818e-16
|
494
|
25
|
18
|
0.72
|
0.0364372469635627
|
|
KEGG
|
KEGG:04514
|
Cell adhesion molecules
|
7.43641128873114e-16
|
146
|
25
|
13
|
0.52
|
0.089041095890411
|
|
KEGG
|
KEGG:05166
|
Human T-cell leukemia virus 1 infection
|
2.66369768871992e-15
|
216
|
25
|
14
|
0.56
|
0.0648148148148148
|
|
KEGG
|
KEGG:05310
|
Asthma
|
3.42399616718515e-14
|
27
|
132
|
12
|
0.0909090909090909
|
0.444444444444444
|
|
KEGG
|
KEGG:04672
|
Intestinal immune network for IgA production
|
9.72271183103954e-14
|
45
|
107
|
13
|
0.121495327102804
|
0.288888888888889
|
|
KEGG
|
KEGG:05321
|
Inflammatory bowel disease
|
8.68353387151345e-13
|
62
|
25
|
9
|
0.36
|
0.145161290322581
|
|
KEGG
|
KEGG:05140
|
Leishmaniasis
|
3.33921794480291e-12
|
72
|
25
|
9
|
0.36
|
0.125
|
|
KEGG
|
KEGG:04658
|
Th1 and Th2 cell differentiation
|
2.26682105395934e-11
|
89
|
25
|
9
|
0.36
|
0.101123595505618
|
|
KEGG
|
KEGG:05150
|
Staphylococcus aureus infection
|
5.36759435210676e-11
|
87
|
109
|
14
|
0.128440366972477
|
0.160919540229885
|
|
KEGG
|
KEGG:04659
|
Th17 cell differentiation
|
8.47518321058457e-11
|
104
|
25
|
9
|
0.36
|
0.0865384615384615
|
|
KEGG
|
KEGG:05145
|
Toxoplasmosis
|
1.23464009528049e-10
|
109
|
25
|
9
|
0.36
|
0.0825688073394495
|
|
KEGG
|
KEGG:05152
|
Tuberculosis
|
2.83873084972289e-10
|
175
|
25
|
10
|
0.4
|
0.0571428571428571
|
|
KEGG
|
KEGG:05323
|
Rheumatoid arthritis
|
8.15357896938181e-10
|
88
|
25
|
8
|
0.32
|
0.0909090909090909
|
|
KEGG
|
KEGG:04640
|
Hematopoietic cell lineage
|
1.45228813331898e-09
|
95
|
25
|
8
|
0.32
|
0.0842105263157895
|
|
KEGG
|
KEGG:05164
|
Influenza A
|
3.0278065448793e-09
|
169
|
32
|
10
|
0.3125
|
0.0591715976331361
|
|
KEGG
|
KEGG:05322
|
Systemic lupus erythematosus
|
1.16115791123941e-08
|
132
|
109
|
14
|
0.128440366972477
|
0.106060606060606
|
|
KEGG
|
KEGG:05170
|
Human immunodeficiency virus 1 infection
|
3.27924263856806e-07
|
210
|
23
|
8
|
0.347826086956522
|
0.0380952380952381
|
|
KEGG
|
KEGG:05163
|
Human cytomegalovirus infection
|
8.1365216002487e-06
|
223
|
15
|
6
|
0.4
|
0.0269058295964126
|
|
KEGG
|
KEGG:03050
|
Proteasome
|
1.25402476433669e-05
|
46
|
40
|
5
|
0.125
|
0.108695652173913
|
|
KEGG
|
KEGG:05167
|
Kaposi sarcoma-associated herpesvirus infection
|
7.25711708413514e-05
|
193
|
24
|
6
|
0.25
|
0.0310880829015544
|
|
KEGG
|
KEGG:04061
|
Viral protein interaction with cytokine and cytokine receptor
|
0.000153847935648785
|
98
|
135
|
9
|
0.0666666666666667
|
0.0918367346938776
|
|
KEGG
|
KEGG:04218
|
Cellular senescence
|
0.000265529951879381
|
156
|
23
|
5
|
0.217391304347826
|
0.032051282051282
|
|
KEGG
|
KEGG:05165
|
Human papillomavirus infection
|
0.00053035693821206
|
331
|
29
|
7
|
0.241379310344828
|
0.0211480362537764
|
|
KEGG
|
KEGG:05203
|
Viral carcinogenesis
|
0.000842994506999791
|
202
|
23
|
5
|
0.217391304347826
|
0.0247524752475248
|
|
KEGG
|
KEGG:05340
|
Primary immunodeficiency
|
0.000938812888087625
|
37
|
126
|
5
|
0.0396825396825397
|
0.135135135135135
|
|
KEGG
|
KEGG:04650
|
Natural killer cell mediated cytotoxicity
|
0.00131456074713706
|
123
|
23
|
4
|
0.173913043478261
|
0.032520325203252
|
|
KEGG
|
KEGG:04144
|
Endocytosis
|
0.00213918803741354
|
252
|
23
|
5
|
0.217391304347826
|
0.0198412698412698
|
|
KEGG
|
KEGG:04620
|
Toll-like receptor signaling pathway
|
0.00312021124756966
|
102
|
123
|
7
|
0.0569105691056911
|
0.0686274509803922
|
|
KEGG
|
KEGG:04060
|
Cytokine-cytokine receptor interaction
|
0.00417217868945516
|
293
|
135
|
13
|
0.0962962962962963
|
0.0443686006825939
|
|
KEGG
|
KEGG:05133
|
Pertussis
|
0.0114956302373419
|
76
|
109
|
5
|
0.0458715596330275
|
0.0657894736842105
|
|
KEGG
|
KEGG:04610
|
Complement and coagulation cascades
|
0.0176092563205241
|
85
|
109
|
5
|
0.0458715596330275
|
0.0588235294117647
|
|
KEGG
|
KEGG:04621
|
NOD-like receptor signaling pathway
|
0.0176092563205241
|
177
|
123
|
8
|
0.0650406504065041
|
0.0451977401129944
|
|
KEGG
|
KEGG:04062
|
Chemokine signaling pathway
|
0.0423475215095323
|
190
|
135
|
8
|
0.0592592592592593
|
0.0421052631578947
|
|
KEGG
|
KEGG:04623
|
Cytosolic DNA-sensing pathway
|
0.0423475215095323
|
62
|
123
|
4
|
0.032520325203252
|
0.0645161290322581
|
|
MIRNA
|
MIRNA:hsa-miR-146a-5p
|
hsa-miR-146a-5p
|
3.8670440478953e-05
|
200
|
91
|
11
|
0.120879120879121
|
0.055
|
|
REAC
|
REAC:R-HSA-913531
|
Interferon Signaling
|
2.44539891206601e-25
|
191
|
79
|
27
|
0.341772151898734
|
0.141361256544503
|
|
REAC
|
REAC:R-HSA-877300
|
Interferon gamma signaling
|
8.70158844202861e-19
|
87
|
35
|
14
|
0.4
|
0.160919540229885
|
|
REAC
|
REAC:R-HSA-1236975
|
Antigen processing-Cross presentation
|
4.89419842034846e-17
|
99
|
41
|
14
|
0.341463414634146
|
0.141414141414141
|
|
REAC
|
REAC:R-HSA-1280215
|
Cytokine Signaling in Immune system
|
4.89419842034846e-17
|
842
|
35
|
24
|
0.685714285714286
|
0.0285035629453682
|
|
REAC
|
REAC:R-HSA-909733
|
Interferon alpha/beta signaling
|
4.89419842034846e-17
|
66
|
79
|
15
|
0.189873417721519
|
0.227272727272727
|
|
REAC
|
REAC:R-HSA-1280218
|
Adaptive Immune System
|
7.46856440177517e-17
|
800
|
25
|
20
|
0.8
|
0.025
|
|
REAC
|
REAC:R-HSA-1236974
|
ER-Phagosome pathway
|
9.19691364874275e-17
|
83
|
40
|
13
|
0.325
|
0.156626506024096
|
|
REAC
|
REAC:R-HSA-168256
|
Immune System
|
6.81954804063536e-15
|
2139
|
58
|
41
|
0.706896551724138
|
0.0191678354371202
|
|
REAC
|
REAC:R-HSA-1236977
|
Endosomal/Vacuolar pathway
|
1.07740335765455e-13
|
11
|
41
|
7
|
0.170731707317073
|
0.636363636363636
|
|
REAC
|
REAC:R-HSA-983170
|
Antigen Presentation: Folding, assembly and peptide loading of class I MHC
|
3.15543646704656e-13
|
25
|
36
|
8
|
0.222222222222222
|
0.32
|
|
REAC
|
REAC:R-HSA-202424
|
Downstream TCR signaling
|
5.52527108785583e-12
|
98
|
22
|
9
|
0.409090909090909
|
0.0918367346938776
|
|
REAC
|
REAC:R-HSA-202403
|
TCR signaling
|
3.2835630534374e-11
|
120
|
22
|
9
|
0.409090909090909
|
0.075
|
|
REAC
|
REAC:R-HSA-983169
|
Class I MHC mediated antigen processing & presentation
|
8.14244744523474e-11
|
370
|
23
|
12
|
0.521739130434783
|
0.0324324324324324
|
|
REAC
|
REAC:R-HSA-2132295
|
MHC class II antigen presentation
|
6.76622558656764e-09
|
123
|
25
|
8
|
0.32
|
0.0650406504065041
|
|
REAC
|
REAC:R-HSA-202430
|
Translocation of ZAP-70 to Immunological synapse
|
9.32211856686034e-09
|
20
|
22
|
5
|
0.227272727272727
|
0.25
|
|
REAC
|
REAC:R-HSA-202427
|
Phosphorylation of CD3 and TCR zeta chains
|
1.88912372850088e-08
|
23
|
22
|
5
|
0.227272727272727
|
0.217391304347826
|
|
REAC
|
REAC:R-HSA-389948
|
PD-1 signaling
|
2.24287165535307e-08
|
24
|
22
|
5
|
0.227272727272727
|
0.208333333333333
|
|
REAC
|
REAC:R-HSA-202433
|
Generation of second messenger molecules
|
1.59407669319924e-07
|
35
|
22
|
5
|
0.227272727272727
|
0.142857142857143
|
|
REAC
|
REAC:R-HSA-388841
|
Costimulation by the CD28 family
|
3.65554142324937e-06
|
67
|
9
|
4
|
0.444444444444444
|
0.0597014925373134
|
|
REAC
|
REAC:R-HSA-8939902
|
Regulation of RUNX2 expression and activity
|
4.56750209526935e-06
|
71
|
40
|
6
|
0.15
|
0.0845070422535211
|
|
REAC
|
REAC:R-HSA-75815
|
Ubiquitin-dependent degradation of Cyclin D
|
1.80529571608705e-05
|
52
|
40
|
5
|
0.125
|
0.0961538461538462
|
|
REAC
|
REAC:R-HSA-349425
|
Autodegradation of the E3 ubiquitin ligase COP1
|
1.80529571608705e-05
|
52
|
40
|
5
|
0.125
|
0.0961538461538462
|
|
REAC
|
REAC:R-HSA-9604323
|
Negative regulation of NOTCH4 signaling
|
1.80529571608705e-05
|
53
|
40
|
5
|
0.125
|
0.0943396226415094
|
|
REAC
|
REAC:R-HSA-169911
|
Regulation of Apoptosis
|
1.80529571608705e-05
|
53
|
40
|
5
|
0.125
|
0.0943396226415094
|
|
REAC
|
REAC:R-HSA-1236978
|
Cross-presentation of soluble exogenous antigens (endosomes)
|
1.80529571608705e-05
|
50
|
40
|
5
|
0.125
|
0.1
|
|
REAC
|
REAC:R-HSA-180585
|
Vif-mediated degradation of APOBEC3G
|
1.80529571608705e-05
|
53
|
40
|
5
|
0.125
|
0.0943396226415094
|
|
REAC
|
REAC:R-HSA-211733
|
Regulation of activated PAK-2p34 by proteasome mediated degradation
|
1.80529571608705e-05
|
50
|
40
|
5
|
0.125
|
0.1
|
|
REAC
|
REAC:R-HSA-69610
|
p53-Independent DNA Damage Response
|
1.80529571608705e-05
|
52
|
40
|
5
|
0.125
|
0.0961538461538462
|
|
REAC
|
REAC:R-HSA-350562
|
Regulation of ornithine decarboxylase (ODC)
|
1.80529571608705e-05
|
51
|
40
|
5
|
0.125
|
0.0980392156862745
|
|
REAC
|
REAC:R-HSA-69601
|
Ubiquitin Mediated Degradation of Phosphorylated Cdc25A
|
1.80529571608705e-05
|
52
|
40
|
5
|
0.125
|
0.0961538461538462
|
|
REAC
|
REAC:R-HSA-180534
|
Vpu mediated degradation of CD4
|
1.80529571608705e-05
|
52
|
40
|
5
|
0.125
|
0.0961538461538462
|
|
REAC
|
REAC:R-HSA-69613
|
p53-Independent G1/S DNA damage checkpoint
|
1.80529571608705e-05
|
52
|
40
|
5
|
0.125
|
0.0961538461538462
|
|
REAC
|
REAC:R-HSA-8854050
|
FBXL7 down-regulates AURKA during mitotic entry and in early mitosis
|
1.86133839307075e-05
|
55
|
40
|
5
|
0.125
|
0.0909090909090909
|
|
REAC
|
REAC:R-HSA-450408
|
AUF1 (hnRNP D0) binds and destabilizes mRNA
|
1.86133839307075e-05
|
54
|
40
|
5
|
0.125
|
0.0925925925925926
|
|
REAC
|
REAC:R-HSA-198933
|
Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell
|
1.86133839307075e-05
|
181
|
23
|
6
|
0.260869565217391
|
0.0331491712707182
|
|
REAC
|
REAC:R-HSA-174113
|
SCF-beta-TrCP mediated degradation of Emi1
|
1.86133839307075e-05
|
55
|
40
|
5
|
0.125
|
0.0909090909090909
|
|
REAC
|
REAC:R-HSA-4641257
|
Degradation of AXIN
|
1.86133839307075e-05
|
55
|
40
|
5
|
0.125
|
0.0909090909090909
|
|
REAC
|
REAC:R-HSA-8941858
|
Regulation of RUNX3 expression and activity
|
1.86133839307075e-05
|
54
|
40
|
5
|
0.125
|
0.0925925925925926
|
|
REAC
|
REAC:R-HSA-5362768
|
Hh mutants are degraded by ERAD
|
1.95537830570516e-05
|
56
|
40
|
5
|
0.125
|
0.0892857142857143
|
|
REAC
|
REAC:R-HSA-4641258
|
Degradation of DVL
|
2.03319644873825e-05
|
57
|
40
|
5
|
0.125
|
0.087719298245614
|
|
REAC
|
REAC:R-HSA-69541
|
Stabilization of p53
|
2.03319644873825e-05
|
57
|
40
|
5
|
0.125
|
0.087719298245614
|
|
REAC
|
REAC:R-HSA-187577
|
SCF(Skp2)-mediated degradation of p27/p21
|
2.11436924384917e-05
|
60
|
40
|
5
|
0.125
|
0.0833333333333333
|
|
REAC
|
REAC:R-HSA-5676590
|
NIK-->noncanonical NF-kB signaling
|
2.11436924384917e-05
|
59
|
40
|
5
|
0.125
|
0.0847457627118644
|
|
REAC
|
REAC:R-HSA-351202
|
Metabolism of polyamines
|
2.11436924384917e-05
|
58
|
40
|
5
|
0.125
|
0.0862068965517241
|
|
REAC
|
REAC:R-HSA-8851680
|
Butyrophilin (BTN) family interactions
|
2.11436924384917e-05
|
12
|
91
|
4
|
0.043956043956044
|
0.333333333333333
|
|
REAC
|
REAC:R-HSA-5610780
|
Degradation of GLI1 by the proteasome
|
2.11436924384917e-05
|
60
|
40
|
5
|
0.125
|
0.0833333333333333
|
|
REAC
|
REAC:R-HSA-5610783
|
Degradation of GLI2 by the proteasome
|
2.11436924384917e-05
|
60
|
40
|
5
|
0.125
|
0.0833333333333333
|
|
REAC
|
REAC:R-HSA-5607761
|
Dectin-1 mediated noncanonical NF-kB signaling
|
2.11436924384917e-05
|
60
|
40
|
5
|
0.125
|
0.0833333333333333
|
|
REAC
|
REAC:R-HSA-5610785
|
GLI3 is processed to GLI3R by the proteasome
|
2.11436924384917e-05
|
60
|
40
|
5
|
0.125
|
0.0833333333333333
|
|
REAC
|
REAC:R-HSA-68827
|
CDT1 association with the CDC6:ORC:origin complex
|
2.11436924384917e-05
|
59
|
40
|
5
|
0.125
|
0.0847457627118644
|
|
REAC
|
REAC:R-HSA-5387390
|
Hh mutants abrogate ligand secretion
|
2.11436924384917e-05
|
59
|
40
|
5
|
0.125
|
0.0847457627118644
|
|
REAC
|
REAC:R-HSA-5678895
|
Defective CFTR causes cystic fibrosis
|
2.2526087563322e-05
|
61
|
40
|
5
|
0.125
|
0.0819672131147541
|
|
REAC
|
REAC:R-HSA-162909
|
Host Interactions of HIV factors
|
2.70223147299298e-05
|
125
|
21
|
5
|
0.238095238095238
|
0.04
|
|
REAC
|
REAC:R-HSA-174084
|
Autodegradation of Cdh1 by Cdh1:APC/C
|
2.70690285661567e-05
|
64
|
40
|
5
|
0.125
|
0.078125
|
|
REAC
|
REAC:R-HSA-4608870
|
Asymmetric localization of PCP proteins
|
2.70690285661567e-05
|
64
|
40
|
5
|
0.125
|
0.078125
|
|
REAC
|
REAC:R-HSA-5358346
|
Hedgehog ligand biogenesis
|
2.87214090487667e-05
|
65
|
40
|
5
|
0.125
|
0.0769230769230769
|
|
REAC
|
REAC:R-HSA-1234176
|
Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha
|
2.94138696513044e-05
|
66
|
40
|
5
|
0.125
|
0.0757575757575758
|
|
REAC
|
REAC:R-HSA-69563
|
p53-Dependent G1 DNA Damage Response
|
2.94138696513044e-05
|
66
|
40
|
5
|
0.125
|
0.0757575757575758
|
|
REAC
|
REAC:R-HSA-69580
|
p53-Dependent G1/S DNA damage checkpoint
|
2.94138696513044e-05
|
66
|
40
|
5
|
0.125
|
0.0757575757575758
|
|
REAC
|
REAC:R-HSA-8878166
|
Transcriptional regulation by RUNX2
|
3.01642065942206e-05
|
118
|
40
|
6
|
0.15
|
0.0508474576271186
|
|
REAC
|
REAC:R-HSA-1169091
|
Activation of NF-kappaB in B cells
|
3.01642065942206e-05
|
67
|
40
|
5
|
0.125
|
0.0746268656716418
|
|
REAC
|
REAC:R-HSA-5658442
|
Regulation of RAS by GAPs
|
3.01642065942206e-05
|
67
|
40
|
5
|
0.125
|
0.0746268656716418
|
|
REAC
|
REAC:R-HSA-174154
|
APC/C:Cdc20 mediated degradation of Securin
|
3.09695512842396e-05
|
68
|
40
|
5
|
0.125
|
0.0735294117647059
|
|
REAC
|
REAC:R-HSA-69615
|
G1/S DNA Damage Checkpoints
|
3.09695512842396e-05
|
68
|
40
|
5
|
0.125
|
0.0735294117647059
|
|
REAC
|
REAC:R-HSA-68867
|
Assembly of the pre-replicative complex
|
3.09695512842396e-05
|
68
|
40
|
5
|
0.125
|
0.0735294117647059
|
|
REAC
|
REAC:R-HSA-8948751
|
Regulation of PTEN stability and activity
|
3.27921403108529e-05
|
69
|
40
|
5
|
0.125
|
0.072463768115942
|
|
REAC
|
REAC:R-HSA-68949
|
Orc1 removal from chromatin
|
3.72160799895067e-05
|
71
|
40
|
5
|
0.125
|
0.0704225352112676
|
|
REAC
|
REAC:R-HSA-380108
|
Chemokine receptors bind chemokines
|
4.08668981110397e-05
|
57
|
135
|
7
|
0.0518518518518519
|
0.12280701754386
|
|
REAC
|
REAC:R-HSA-69017
|
CDK-mediated phosphorylation and removal of Cdc6
|
4.08668981110397e-05
|
73
|
40
|
5
|
0.125
|
0.0684931506849315
|
|
REAC
|
REAC:R-HSA-174184
|
Cdc20:Phospho-APC/C mediated degradation of Cyclin A
|
4.08668981110397e-05
|
73
|
40
|
5
|
0.125
|
0.0684931506849315
|
|
REAC
|
REAC:R-HSA-174178
|
APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1
|
4.24928807360587e-05
|
74
|
40
|
5
|
0.125
|
0.0675675675675676
|
|
REAC
|
REAC:R-HSA-179419
|
APC:Cdc20 mediated degradation of cell cycle proteins prior to satisfation of the cell cycle checkpoint
|
4.24928807360587e-05
|
74
|
40
|
5
|
0.125
|
0.0675675675675676
|
|
REAC
|
REAC:R-HSA-1234174
|
Cellular response to hypoxia
|
4.47801314271766e-05
|
75
|
40
|
5
|
0.125
|
0.0666666666666667
|
|
REAC
|
REAC:R-HSA-5668541
|
TNFR2 non-canonical NF-kB pathway
|
4.5872458988899e-05
|
100
|
52
|
6
|
0.115384615384615
|
0.06
|
|
REAC
|
REAC:R-HSA-176409
|
APC/C:Cdc20 mediated degradation of mitotic proteins
|
4.65263021883085e-05
|
76
|
40
|
5
|
0.125
|
0.0657894736842105
|
|
REAC
|
REAC:R-HSA-176814
|
Activation of APC/C and APC/C:Cdc20 mediated degradation of mitotic proteins
|
4.83307617525144e-05
|
77
|
40
|
5
|
0.125
|
0.0649350649350649
|
|
REAC
|
REAC:R-HSA-5619084
|
ABC transporter disorders
|
4.83307617525144e-05
|
77
|
40
|
5
|
0.125
|
0.0649350649350649
|
|
REAC
|
REAC:R-HSA-8852276
|
The role of GTSE1 in G2/M progression after G2 checkpoint
|
5.41227131009581e-05
|
79
|
40
|
5
|
0.125
|
0.0632911392405063
|
|
REAC
|
REAC:R-HSA-1168372
|
Downstream signaling events of B Cell Receptor (BCR)
|
5.68423651775045e-05
|
80
|
40
|
5
|
0.125
|
0.0625
|
|
REAC
|
REAC:R-HSA-9013694
|
Signaling by NOTCH4
|
5.89226118137143e-05
|
81
|
40
|
5
|
0.125
|
0.0617283950617284
|
|
REAC
|
REAC:R-HSA-176408
|
Regulation of APC/C activators between G1/S and early anaphase
|
5.89226118137143e-05
|
81
|
40
|
5
|
0.125
|
0.0617283950617284
|
|
REAC
|
REAC:R-HSA-69202
|
Cyclin E associated events during G1/S transition
|
6.40305230591604e-05
|
83
|
40
|
5
|
0.125
|
0.0602409638554217
|
|
REAC
|
REAC:R-HSA-195253
|
Degradation of beta-catenin by the destruction complex
|
6.40305230591604e-05
|
83
|
40
|
5
|
0.125
|
0.0602409638554217
|
|
REAC
|
REAC:R-HSA-168249
|
Innate Immune System
|
6.40305230591604e-05
|
1073
|
21
|
10
|
0.476190476190476
|
0.00931966449207828
|
|
REAC
|
REAC:R-HSA-69656
|
Cyclin A:Cdk2-associated events at S phase entry
|
6.94577870305714e-05
|
85
|
40
|
5
|
0.125
|
0.0588235294117647
|
|
REAC
|
REAC:R-HSA-69002
|
DNA Replication Pre-Initiation
|
6.94577870305714e-05
|
85
|
40
|
5
|
0.125
|
0.0588235294117647
|
|
REAC
|
REAC:R-HSA-5632684
|
Hedgehog 'on' state
|
6.94577870305714e-05
|
85
|
40
|
5
|
0.125
|
0.0588235294117647
|
|
REAC
|
REAC:R-HSA-450531
|
Regulation of mRNA stability by proteins that bind AU-rich elements
|
7.27055097902366e-05
|
86
|
40
|
5
|
0.125
|
0.0581395348837209
|
|
REAC
|
REAC:R-HSA-174143
|
APC/C-mediated degradation of cell cycle proteins
|
7.95261973345654e-05
|
88
|
40
|
5
|
0.125
|
0.0568181818181818
|
|
REAC
|
REAC:R-HSA-453276
|
Regulation of mitotic cell cycle
|
7.95261973345654e-05
|
88
|
40
|
5
|
0.125
|
0.0568181818181818
|
|
REAC
|
REAC:R-HSA-5687128
|
MAPK6/MAPK4 signaling
|
8.31033316905083e-05
|
89
|
40
|
5
|
0.125
|
0.0561797752808989
|
|
REAC
|
REAC:R-HSA-69052
|
Switching of origins to a post-replicative state
|
9.15864658168985e-05
|
91
|
40
|
5
|
0.125
|
0.0549450549450549
|
|
REAC
|
REAC:R-HSA-4086400
|
PCP/CE pathway
|
9.45270802019694e-05
|
92
|
40
|
5
|
0.125
|
0.0543478260869565
|
|
REAC
|
REAC:R-HSA-5689603
|
UCH proteinases
|
9.45270802019694e-05
|
92
|
40
|
5
|
0.125
|
0.0543478260869565
|
|
REAC
|
REAC:R-HSA-8939236
|
RUNX1 regulates transcription of genes involved in differentiation of HSCs
|
0.000102741000304565
|
94
|
40
|
5
|
0.125
|
0.0531914893617021
|
|
REAC
|
REAC:R-HSA-8878159
|
Transcriptional regulation by RUNX3
|
0.000102741000304565
|
94
|
40
|
5
|
0.125
|
0.0531914893617021
|
|
REAC
|
REAC:R-HSA-5607764
|
CLEC7A (Dectin-1) signaling
|
0.000118393366910053
|
97
|
40
|
5
|
0.125
|
0.0515463917525773
|
|
REAC
|
REAC:R-HSA-9020702
|
Interleukin-1 signaling
|
0.000142441243373795
|
101
|
40
|
5
|
0.125
|
0.0495049504950495
|
|
REAC
|
REAC:R-HSA-382556
|
ABC-family proteins mediated transport
|
0.000154974097676177
|
103
|
40
|
5
|
0.125
|
0.0485436893203883
|
|
REAC
|
REAC:R-HSA-5610787
|
Hedgehog 'off' state
|
0.000239278557058356
|
113
|
40
|
5
|
0.125
|
0.0442477876106195
|
|
REAC
|
REAC:R-HSA-9683610
|
Maturation of nucleoprotein
|
0.000248428317842978
|
10
|
89
|
3
|
0.0337078651685393
|
0.3
|
|
REAC
|
REAC:R-HSA-162906
|
HIV Infection
|
0.000248428317842978
|
226
|
21
|
5
|
0.238095238095238
|
0.0221238938053097
|
|
REAC
|
REAC:R-HSA-157118
|
Signaling by NOTCH
|
0.000253579518371989
|
197
|
24
|
5
|
0.208333333333333
|
0.0253807106598985
|
|
REAC
|
REAC:R-HSA-69239
|
Synthesis of DNA
|
0.000299718352127985
|
120
|
21
|
4
|
0.19047619047619
|
0.0333333333333333
|
|
REAC
|
REAC:R-HSA-69306
|
DNA Replication
|
0.000381554382594412
|
128
|
21
|
4
|
0.19047619047619
|
0.03125
|
|
REAC
|
REAC:R-HSA-69206
|
G1/S Transition
|
0.000413497216728008
|
131
|
21
|
4
|
0.19047619047619
|
0.0305343511450382
|
|
REAC
|
REAC:R-HSA-5621481
|
C-type lectin receptors (CLRs)
|
0.000469198873124197
|
136
|
21
|
4
|
0.19047619047619
|
0.0294117647058824
|
|
REAC
|
REAC:R-HSA-2871837
|
FCERI mediated NF-kB activation
|
0.000469198873124197
|
136
|
21
|
4
|
0.19047619047619
|
0.0294117647058824
|
|
REAC
|
REAC:R-HSA-196807
|
Nicotinate metabolism
|
0.000487021721864988
|
31
|
92
|
4
|
0.0434782608695652
|
0.129032258064516
|
|
REAC
|
REAC:R-HSA-446652
|
Interleukin-1 family signaling
|
0.000487021721864988
|
138
|
21
|
4
|
0.19047619047619
|
0.0289855072463768
|
|
REAC
|
REAC:R-HSA-109581
|
Apoptosis
|
0.000487021721864988
|
174
|
49
|
6
|
0.122448979591837
|
0.0344827586206897
|
|
REAC
|
REAC:R-HSA-6807070
|
PTEN Regulation
|
0.000487021721864988
|
138
|
21
|
4
|
0.19047619047619
|
0.0289855072463768
|
|
REAC
|
REAC:R-HSA-5357801
|
Programmed Cell Death
|
0.000530043834808989
|
177
|
49
|
6
|
0.122448979591837
|
0.0338983050847458
|
|
REAC
|
REAC:R-HSA-3858494
|
Beta-catenin independent WNT signaling
|
0.000539610290059056
|
143
|
21
|
4
|
0.19047619047619
|
0.027972027972028
|
|
REAC
|
REAC:R-HSA-453279
|
Mitotic G1 phase and G1/S transition
|
0.000610664043071788
|
148
|
21
|
4
|
0.19047619047619
|
0.027027027027027
|
|
REAC
|
REAC:R-HSA-5358351
|
Signaling by Hedgehog
|
0.000615984670710434
|
149
|
21
|
4
|
0.19047619047619
|
0.0268456375838926
|
|
REAC
|
REAC:R-HSA-69481
|
G2/M Checkpoints
|
0.000615984670710434
|
149
|
21
|
4
|
0.19047619047619
|
0.0268456375838926
|
|
REAC
|
REAC:R-HSA-8951664
|
Neddylation
|
0.000717751573402303
|
233
|
40
|
6
|
0.15
|
0.0257510729613734
|
|
REAC
|
REAC:R-HSA-69242
|
S Phase
|
0.000834896373919167
|
162
|
21
|
4
|
0.19047619047619
|
0.0246913580246914
|
|
REAC
|
REAC:R-HSA-983705
|
Signaling by the B Cell Receptor (BCR)
|
0.00086773458949971
|
164
|
21
|
4
|
0.19047619047619
|
0.024390243902439
|
|
REAC
|
REAC:R-HSA-983168
|
Antigen processing: Ubiquitination & Proteasome degradation
|
0.000893024641095733
|
308
|
21
|
5
|
0.238095238095238
|
0.0162337662337662
|
|
REAC
|
REAC:R-HSA-449147
|
Signaling by Interleukins
|
0.000993016047087905
|
446
|
42
|
8
|
0.19047619047619
|
0.0179372197309417
|
|
REAC
|
REAC:R-HSA-9010553
|
Regulation of expression of SLITs and ROBOs
|
0.000993096164533696
|
171
|
21
|
4
|
0.19047619047619
|
0.0233918128654971
|
|
REAC
|
REAC:R-HSA-5619115
|
Disorders of transmembrane transporters
|
0.00102973946058989
|
173
|
21
|
4
|
0.19047619047619
|
0.023121387283237
|
|
REAC
|
REAC:R-HSA-2454202
|
Fc epsilon receptor (FCERI) signaling
|
0.00122904308664657
|
182
|
21
|
4
|
0.19047619047619
|
0.021978021978022
|
|
REAC
|
REAC:R-HSA-2467813
|
Separation of Sister Chromatids
|
0.00137899863422082
|
188
|
21
|
4
|
0.19047619047619
|
0.0212765957446809
|
|
REAC
|
REAC:R-HSA-197264
|
Nicotinamide salvaging
|
0.00151297163830571
|
19
|
89
|
3
|
0.0337078651685393
|
0.157894736842105
|
|
REAC
|
REAC:R-HSA-201681
|
TCF dependent signaling in response to WNT
|
0.00157719062224088
|
196
|
21
|
4
|
0.19047619047619
|
0.0204081632653061
|
|
REAC
|
REAC:R-HSA-69275
|
G2/M Transition
|
0.00157719062224088
|
196
|
21
|
4
|
0.19047619047619
|
0.0204081632653061
|
|
REAC
|
REAC:R-HSA-453274
|
Mitotic G2-G2/M phases
|
0.00162630539676266
|
198
|
21
|
4
|
0.19047619047619
|
0.0202020202020202
|
|
REAC
|
REAC:R-HSA-8878171
|
Transcriptional regulation by RUNX1
|
0.00166374814659151
|
200
|
21
|
4
|
0.19047619047619
|
0.02
|
|
REAC
|
REAC:R-HSA-5689880
|
Ub-specific processing proteases
|
0.00166374814659151
|
200
|
21
|
4
|
0.19047619047619
|
0.02
|
|
REAC
|
REAC:R-HSA-376176
|
Signaling by ROBO receptors
|
0.00222730190829495
|
217
|
21
|
4
|
0.19047619047619
|
0.0184331797235023
|
|
REAC
|
REAC:R-HSA-68882
|
Mitotic Anaphase
|
0.00265987308955247
|
228
|
21
|
4
|
0.19047619047619
|
0.0175438596491228
|
|
REAC
|
REAC:R-HSA-2555396
|
Mitotic Metaphase and Anaphase
|
0.00268391410131649
|
229
|
21
|
4
|
0.19047619047619
|
0.0174672489082969
|
|
REAC
|
REAC:R-HSA-5663202
|
Diseases of signal transduction by growth factor receptors and second messengers
|
0.00352632499991936
|
371
|
24
|
5
|
0.208333333333333
|
0.0134770889487871
|
|
REAC
|
REAC:R-HSA-1257604
|
PIP3 activates AKT signaling
|
0.00361059188227564
|
249
|
21
|
4
|
0.19047619047619
|
0.0160642570281124
|
|
REAC
|
REAC:R-HSA-9683701
|
Translation of structural proteins
|
0.00441706580088292
|
28
|
89
|
3
|
0.0337078651685393
|
0.107142857142857
|
|
REAC
|
REAC:R-HSA-5673001
|
RAF/MAP kinase cascade
|
0.00441706580088292
|
264
|
21
|
4
|
0.19047619047619
|
0.0151515151515152
|
|
REAC
|
REAC:R-HSA-5684996
|
MAPK1/MAPK3 signaling
|
0.0046991908345857
|
269
|
21
|
4
|
0.19047619047619
|
0.0148698884758364
|
|
REAC
|
REAC:R-HSA-69620
|
Cell Cycle Checkpoints
|
0.0047949872107715
|
271
|
21
|
4
|
0.19047619047619
|
0.014760147601476
|
|
REAC
|
REAC:R-HSA-5688426
|
Deubiquitination
|
0.00506335302504183
|
276
|
21
|
4
|
0.19047619047619
|
0.0144927536231884
|
|
REAC
|
REAC:R-HSA-9607240
|
FLT3 Signaling
|
0.00515665648318284
|
278
|
21
|
4
|
0.19047619047619
|
0.0143884892086331
|
|
REAC
|
REAC:R-HSA-9006925
|
Intracellular signaling by second messengers
|
0.00590159812731934
|
289
|
21
|
4
|
0.19047619047619
|
0.013840830449827
|
|
REAC
|
REAC:R-HSA-195721
|
Signaling by WNT
|
0.00608647603104174
|
292
|
21
|
4
|
0.19047619047619
|
0.0136986301369863
|
|
REAC
|
REAC:R-HSA-71291
|
Metabolism of amino acids and derivatives
|
0.00620517852593789
|
368
|
40
|
6
|
0.15
|
0.016304347826087
|
|
REAC
|
REAC:R-HSA-5683057
|
MAPK family signaling cascades
|
0.00728862369801189
|
308
|
21
|
4
|
0.19047619047619
|
0.012987012987013
|
|
REAC
|
REAC:R-HSA-6798695
|
Neutrophil degranulation
|
0.0106441471637237
|
474
|
8
|
3
|
0.375
|
0.00632911392405063
|
|
REAC
|
REAC:R-HSA-451927
|
Interleukin-2 family signaling
|
0.0114261580753921
|
44
|
81
|
3
|
0.037037037037037
|
0.0681818181818182
|
|
REAC
|
REAC:R-HSA-68886
|
M Phase
|
0.0137267477365132
|
371
|
21
|
4
|
0.19047619047619
|
0.0107816711590297
|
|
REAC
|
REAC:R-HSA-9678108
|
SARS-CoV-1 Infection
|
0.020538297292683
|
50
|
89
|
3
|
0.0337078651685393
|
0.06
|
|
REAC
|
REAC:R-HSA-375276
|
Peptide ligand-binding receptors
|
0.0291567676529731
|
195
|
135
|
7
|
0.0518518518518519
|
0.0358974358974359
|
|
REAC
|
REAC:R-HSA-6783783
|
Interleukin-10 signaling
|
0.0343315856884118
|
45
|
123
|
3
|
0.024390243902439
|
0.0666666666666667
|
|
REAC
|
REAC:R-HSA-69278
|
Cell Cycle, Mitotic
|
0.0379774694665152
|
513
|
21
|
4
|
0.19047619047619
|
0.00779727095516569
|
|
REAC
|
REAC:R-HSA-5663205
|
Infectious disease
|
0.0423982182747049
|
815
|
21
|
5
|
0.238095238095238
|
0.00613496932515337
|
|
REAC
|
REAC:R-HSA-977606
|
Regulation of Complement cascade
|
0.0439526177765537
|
101
|
109
|
4
|
0.036697247706422
|
0.0396039603960396
|
|
REAC
|
REAC:R-HSA-196849
|
Metabolism of water-soluble vitamins and cofactors
|
0.0440870172804559
|
120
|
92
|
4
|
0.0434782608695652
|
0.0333333333333333
|
|
REAC
|
REAC:R-HSA-2262752
|
Cellular responses to stress
|
0.0446269325179089
|
546
|
21
|
4
|
0.19047619047619
|
0.00732600732600733
|
|
REAC
|
REAC:R-HSA-422475
|
Axon guidance
|
0.0451831884296367
|
549
|
21
|
4
|
0.19047619047619
|
0.00728597449908925
|
|
REAC
|
REAC:R-HSA-8953897
|
Cellular responses to external stimuli
|
0.0466778901505786
|
560
|
21
|
4
|
0.19047619047619
|
0.00714285714285714
|
|
REAC
|
REAC:R-HSA-9675108
|
Nervous system development
|
0.0498016292845307
|
574
|
21
|
4
|
0.19047619047619
|
0.00696864111498258
|
|
TF
|
TF:M09957
|
Factor: IRF-2; motif: NAAANNGAAAGTGAAASTRN
|
4.22201714108179e-29
|
272
|
116
|
32
|
0.275862068965517
|
0.117647058823529
|
|
TF
|
TF:M07216
|
Factor: IRF1; motif: NNNYASTTTCACTTTCNNTTT
|
6.90098597196173e-28
|
440
|
116
|
36
|
0.310344827586207
|
0.0818181818181818
|
|
TF
|
TF:M04788
|
Factor: IRF-1; motif: STTTCACTTTCNNT
|
3.95986837494792e-23
|
536
|
89
|
31
|
0.348314606741573
|
0.0578358208955224
|
|
TF
|
TF:M08887
|
Factor: IRF; motif: NNGAAANTGAAANN
|
3.95986837494792e-23
|
1267
|
130
|
50
|
0.384615384615385
|
0.0394632991318074
|
|
TF
|
TF:M11664
|
Factor: IRF-2; motif: NGAAASYGAAAS
|
9.72342035917312e-23
|
1651
|
116
|
52
|
0.448275862068966
|
0.031496062992126
|
|
TF
|
TF:M09629
|
Factor: IRF-1; motif: NAAANNGAAAGTGAAASTRN
|
3.25493147920604e-20
|
336
|
116
|
27
|
0.232758620689655
|
0.0803571428571429
|
|
TF
|
TF:M08775_1
|
Factor: IRF-2; motif: NAANYGAAASYR; match class: 1
|
6.70504938255325e-20
|
2761
|
129
|
65
|
0.503875968992248
|
0.0235421948569359
|
|
TF
|
TF:M11685
|
Factor: IRF-8; motif: NCGAAACYGAAACYN
|
1.1805248872316e-19
|
1617
|
89
|
42
|
0.471910112359551
|
0.025974025974026
|
|
TF
|
TF:M11665
|
Factor: IRF-2; motif: NGAAASYGAAAS
|
4.2426096531198e-19
|
2396
|
126
|
59
|
0.468253968253968
|
0.0246243739565943
|
|
TF
|
TF:M09956
|
Factor: IRF-1; motif: NAAANNGAAASTGAAASNRN
|
1.56252428874025e-18
|
317
|
116
|
25
|
0.21551724137931
|
0.0788643533123028
|
|
TF
|
TF:M00772
|
Factor: IRF; motif: RRAANTGAAASYGNV
|
5.37236768560603e-18
|
1029
|
122
|
39
|
0.319672131147541
|
0.0379008746355685
|
|
TF
|
TF:M11682
|
Factor: IRF-8; motif: NYGAAASYGAAACYN
|
4.03242813011298e-17
|
3795
|
93
|
58
|
0.623655913978495
|
0.0152832674571805
|
|
TF
|
TF:M11686
|
Factor: IRF-4; motif: NYGAAASYGAAACYN
|
2.70230493979318e-16
|
3391
|
131
|
67
|
0.511450381679389
|
0.0197581834267178
|
|
TF
|
TF:M11684
|
Factor: IRF-8; motif: NYGAAACYGAAACTN
|
6.6759287977492e-16
|
4664
|
131
|
78
|
0.595419847328244
|
0.0167238421955403
|
|
TF
|
TF:M08887_1
|
Factor: IRF; motif: NNGAAANTGAAANN; match class: 1
|
3.18886754311896e-15
|
73
|
116
|
14
|
0.120689655172414
|
0.191780821917808
|
|
TF
|
TF:M00453
|
Factor: IRF-7; motif: TNSGAAWNCGAAANTNNN
|
8.31106406870599e-15
|
3082
|
90
|
49
|
0.544444444444444
|
0.0158987670343932
|
|
TF
|
TF:M07045
|
Factor: IRF-1; motif: NNRAAANNGAAASN
|
1.94773624226859e-14
|
535
|
126
|
27
|
0.214285714285714
|
0.0504672897196262
|
|
TF
|
TF:M11677
|
Factor: IRF-3; motif: NGGAAACNGAAACCGAAACN
|
4.98342135056075e-14
|
470
|
123
|
25
|
0.203252032520325
|
0.0531914893617021
|
|
TF
|
TF:M11679
|
Factor: IRF-7; motif: NCGAAANCGAAANYN
|
5.5540188442756e-14
|
622
|
87
|
24
|
0.275862068965517
|
0.0385852090032154
|
|
TF
|
TF:M08775
|
Factor: IRF-2; motif: NAANYGAAASYR
|
1.33500566410538e-13
|
8370
|
129
|
100
|
0.775193798449612
|
0.01194743130227
|
|
TF
|
TF:M00972
|
Factor: IRF; motif: RAAANTGAAAN
|
1.83338036535268e-13
|
1979
|
138
|
49
|
0.355072463768116
|
0.0247599797877716
|
|
TF
|
TF:M11685_1
|
Factor: IRF-8; motif: NCGAAACYGAAACYN; match class: 1
|
3.17266517957213e-13
|
118
|
126
|
15
|
0.119047619047619
|
0.127118644067797
|
|
TF
|
TF:M11687
|
Factor: IRF-4; motif: NCGAAACCGAAACYN
|
5.92733990666149e-13
|
3595
|
90
|
50
|
0.555555555555556
|
0.0139082058414465
|
|
TF
|
TF:M09733
|
Factor: STAT2; motif: NRGAAANNGAAACTNA
|
7.4045245131213e-13
|
2842
|
116
|
52
|
0.448275862068966
|
0.0182969739619986
|
|
TF
|
TF:M11683
|
Factor: IRF-8; motif: NCGAAACCGAAACYN
|
3.31773231563916e-12
|
4690
|
93
|
57
|
0.612903225806452
|
0.0121535181236674
|
|
TF
|
TF:M04020
|
Factor: IRF8; motif: NCGAAACCGAAACT
|
1.19785562028075e-11
|
992
|
79
|
25
|
0.316455696202532
|
0.0252016129032258
|
|
TF
|
TF:M04018
|
Factor: IRF7; motif: NCGAAARYGAAANT
|
7.23847615319414e-11
|
619
|
79
|
20
|
0.253164556962025
|
0.0323101777059774
|
|
TF
|
TF:M11665_1
|
Factor: IRF-2; motif: NGAAASYGAAAS; match class: 1
|
7.93088809545384e-11
|
246
|
126
|
17
|
0.134920634920635
|
0.0691056910569106
|
|
TF
|
TF:M00453_1
|
Factor: IRF-7; motif: TNSGAAWNCGAAANTNNN; match class: 1
|
8.17480235383737e-11
|
332
|
81
|
16
|
0.197530864197531
|
0.0481927710843374
|
|
TF
|
TF:M11676
|
Factor: IRF-3; motif: NGGAAANGGAAASNGAAACN
|
1.42574298527175e-10
|
562
|
123
|
23
|
0.186991869918699
|
0.0409252669039146
|
|
TF
|
TF:M11670
|
Factor: IRF-5; motif: NYGAAACCGAAACY
|
2.11315746668765e-10
|
871
|
126
|
28
|
0.222222222222222
|
0.0321469575200918
|
|
TF
|
TF:M11664_1
|
Factor: IRF-2; motif: NGAAASYGAAAS; match class: 1
|
3.08872847093782e-10
|
140
|
89
|
12
|
0.134831460674157
|
0.0857142857142857
|
|
TF
|
TF:M04014
|
Factor: IRF3; motif: NNRRAANGGAAACCGAAACYR
|
3.51531243849604e-10
|
278
|
89
|
15
|
0.168539325842697
|
0.0539568345323741
|
|
TF
|
TF:M04015
|
Factor: IRF-4; motif: NCGAAACCGAAACYA
|
4.4027889701654e-10
|
2038
|
79
|
32
|
0.40506329113924
|
0.0157016683022571
|
|
TF
|
TF:M11684_1
|
Factor: IRF-8; motif: NYGAAACYGAAACTN; match class: 1
|
1.87399215945969e-09
|
832
|
80
|
21
|
0.2625
|
0.0252403846153846
|
|
TF
|
TF:M00258
|
Factor: ISGF-3; motif: CAGTTTCWCTTTYCC
|
1.99230157595007e-09
|
823
|
126
|
26
|
0.206349206349206
|
0.031591737545565
|
|
TF
|
TF:M11682_1
|
Factor: IRF-8; motif: NYGAAASYGAAACYN; match class: 1
|
5.47693227495614e-09
|
543
|
89
|
18
|
0.202247191011236
|
0.0331491712707182
|
|
TF
|
TF:M11683_1
|
Factor: IRF-8; motif: NCGAAACCGAAACYN; match class: 1
|
8.81882322076593e-09
|
814
|
116
|
24
|
0.206896551724138
|
0.0294840294840295
|
|
TF
|
TF:M07323
|
Factor: IRF-4; motif: KRAAMNGAAANYN
|
1.24694468719227e-08
|
938
|
87
|
22
|
0.252873563218391
|
0.023454157782516
|
|
TF
|
TF:M11686_1
|
Factor: IRF-4; motif: NYGAAASYGAAACYN; match class: 1
|
1.30540632753364e-08
|
452
|
126
|
19
|
0.150793650793651
|
0.0420353982300885
|
|
TF
|
TF:M00699_1
|
Factor: ICSBP; motif: RAARTGAAACTG; match class: 1
|
2.07992049209437e-08
|
47
|
105
|
8
|
0.0761904761904762
|
0.170212765957447
|
|
TF
|
TF:M09733_1
|
Factor: STAT2; motif: NRGAAANNGAAACTNA; match class: 1
|
3.02843887915742e-08
|
367
|
126
|
17
|
0.134920634920635
|
0.0463215258855586
|
|
TF
|
TF:M01882
|
Factor: IRF-2; motif: NNRAAAGTGAAASNNA
|
3.05400808340402e-08
|
258
|
111
|
14
|
0.126126126126126
|
0.0542635658914729
|
|
TF
|
TF:M11671
|
Factor: IRF-5; motif: NCGAAACCGAAACY
|
3.05400808340402e-08
|
389
|
75
|
14
|
0.186666666666667
|
0.0359897172236504
|
|
TF
|
TF:M11687_1
|
Factor: IRF-4; motif: NCGAAACCGAAACYN; match class: 1
|
1.93539374135409e-07
|
474
|
126
|
18
|
0.142857142857143
|
0.0379746835443038
|
|
TF
|
TF:M00258_1
|
Factor: ISGF-3; motif: CAGTTTCWCTTTYCC; match class: 1
|
1.93700537683801e-07
|
30
|
75
|
6
|
0.08
|
0.2
|
|
TF
|
TF:M00699
|
Factor: ICSBP; motif: RAARTGAAACTG
|
2.94221424318874e-07
|
1184
|
105
|
25
|
0.238095238095238
|
0.0211148648648649
|
|
TF
|
TF:M00287
|
Factor: NF-Y; motif: NNNRRCCAATSRGNNN
|
5.89585357805016e-07
|
1635
|
23
|
13
|
0.565217391304348
|
0.00795107033639144
|
|
TF
|
TF:M04849_1
|
Factor: RelA-p65; motif: AAASTCCC; match class: 1
|
6.15559537674092e-07
|
1170
|
59
|
18
|
0.305084745762712
|
0.0153846153846154
|
|
TF
|
TF:M04683
|
Factor: NF-YA; motif: YTCTSATTGGYYRN
|
6.66345528752083e-07
|
1657
|
23
|
13
|
0.565217391304348
|
0.00784550392275196
|
|
TF
|
TF:M00972_1
|
Factor: IRF; motif: RAAANTGAAAN; match class: 1
|
1.58256475046003e-06
|
205
|
105
|
11
|
0.104761904761905
|
0.0536585365853659
|
|
TF
|
TF:M00063
|
Factor: IRF-2; motif: GAAAAGYGAAASY
|
1.58256475046003e-06
|
685
|
138
|
21
|
0.152173913043478
|
0.0306569343065693
|
|
TF
|
TF:M06507
|
Factor: ZFP90; motif: NGGGASTTTAKA
|
2.4041253702916e-06
|
40
|
15
|
4
|
0.266666666666667
|
0.1
|
|
TF
|
TF:M00687
|
Factor: alpha-CP1; motif: CAGCCAATGAG
|
2.44829116856985e-06
|
2326
|
36
|
18
|
0.5
|
0.00773860705073087
|
|
TF
|
TF:M00772_1
|
Factor: IRF; motif: RRAANTGAAASYGNV; match class: 1
|
2.50234485878791e-06
|
57
|
105
|
7
|
0.0666666666666667
|
0.12280701754386
|
|
TF
|
TF:M09745_1
|
Factor: HELIOS; motif: RNARRRGGAASTGARAN; match class: 1
|
3.93635006935338e-06
|
495
|
116
|
16
|
0.137931034482759
|
0.0323232323232323
|
|
TF
|
TF:M07302
|
Factor: NF-Y; motif: RGCCAATCRGN
|
4.39226079734251e-06
|
1576
|
23
|
12
|
0.521739130434783
|
0.00761421319796954
|
|
TF
|
TF:M09957_1
|
Factor: IRF-2; motif: NAAANNGAAAGTGAAASTRN; match class: 1
|
4.66941068217365e-06
|
22
|
89
|
5
|
0.0561797752808989
|
0.227272727272727
|
|
TF
|
TF:M09745
|
Factor: HELIOS; motif: RNARRRGGAASTGARAN
|
4.83283725633499e-06
|
3582
|
134
|
51
|
0.380597014925373
|
0.0142378559463987
|
|
TF
|
TF:M11680
|
Factor: IRF-9; motif: NYGAAACYGAAACYN
|
4.88170601815985e-06
|
128
|
79
|
8
|
0.10126582278481
|
0.0625
|
|
TF
|
TF:M04015_1
|
Factor: IRF-4; motif: NCGAAACCGAAACYA; match class: 1
|
5.1974525440792e-06
|
158
|
26
|
6
|
0.230769230769231
|
0.0379746835443038
|
|
TF
|
TF:M10080
|
Factor: STAT2; motif: RRGRAANNGAAACTGAAAN
|
5.73360846628156e-06
|
356
|
105
|
13
|
0.123809523809524
|
0.0365168539325843
|
|
TF
|
TF:M09959
|
Factor: IRF-4; motif: NAANGRGGAASTGAAASN
|
5.79880680803199e-06
|
3386
|
134
|
49
|
0.365671641791045
|
0.0144713526284702
|
|
TF
|
TF:M11681
|
Factor: IRF-9; motif: NCGAAACYGAAACYN
|
8.49856484467838e-06
|
97
|
75
|
7
|
0.0933333333333333
|
0.0721649484536082
|
|
TF
|
TF:M01279
|
Factor: IRF-3; motif: NBNBTTTCSCTTT
|
8.49856484467838e-06
|
1647
|
92
|
25
|
0.271739130434783
|
0.0151791135397693
|
|
TF
|
TF:M03916
|
Factor: PRDM1; motif: NRAAAGTGAAAGTNN
|
1.32585117511768e-05
|
948
|
61
|
15
|
0.245901639344262
|
0.0158227848101266
|
|
TF
|
TF:M10006
|
Factor: NF-YC; motif: NNRRCCAATCAGNR
|
1.54750914472339e-05
|
2510
|
24
|
14
|
0.583333333333333
|
0.00557768924302789
|
|
TF
|
TF:M10005
|
Factor: NF-YA; motif: NYRRCCAATCAGAR
|
1.70730139520397e-05
|
2225
|
23
|
13
|
0.565217391304348
|
0.00584269662921348
|
|
TF
|
TF:M04706
|
Factor: RelA; motif: GGGRAWTYC
|
1.74768639786544e-05
|
2278
|
81
|
27
|
0.333333333333333
|
0.0118525021949078
|
|
TF
|
TF:M03882_1
|
Factor: RelB:p50; motif: RGAAANTCCCYNNHGC; match class: 1
|
2.48607219645885e-05
|
714
|
62
|
13
|
0.209677419354839
|
0.0182072829131653
|
|
TF
|
TF:M03793
|
Factor: IRF-7; motif: AARGGAAANNGAA
|
2.60925973782987e-05
|
1987
|
116
|
31
|
0.267241379310345
|
0.015601409159537
|
|
TF
|
TF:M09958
|
Factor: IRF-3; motif: RAAARGGAAANNGAAASNGA
|
2.83625847188221e-05
|
589
|
101
|
15
|
0.148514851485149
|
0.0254668930390492
|
|
TF
|
TF:M11672
|
Factor: IRF-5; motif: NCGAAACCGAAACY
|
3.26895330365967e-05
|
120
|
75
|
7
|
0.0933333333333333
|
0.0583333333333333
|
|
TF
|
TF:M09816
|
Factor: NFYA; motif: RRCCAATCAGN
|
3.96263846763425e-05
|
1969
|
23
|
12
|
0.521739130434783
|
0.00609446419502285
|
|
TF
|
TF:M03916_1
|
Factor: PRDM1; motif: NRAAAGTGAAAGTNN; match class: 1
|
3.97631929992355e-05
|
50
|
61
|
5
|
0.0819672131147541
|
0.1
|
|
TF
|
TF:M07216_1
|
Factor: IRF1; motif: NNNYASTTTCACTTTCNNTTT; match class: 1
|
4.1946793118454e-05
|
35
|
89
|
5
|
0.0561797752808989
|
0.142857142857143
|
|
TF
|
TF:M00208_1
|
Factor: NF-kappaB; motif: NGGGACTTTCCA; match class: 1
|
4.2823333177136e-05
|
604
|
25
|
8
|
0.32
|
0.0132450331125828
|
|
TF
|
TF:M03793_1
|
Factor: IRF-7; motif: AARGGAAANNGAA; match class: 1
|
4.53680151949136e-05
|
138
|
101
|
8
|
0.0792079207920792
|
0.0579710144927536
|
|
TF
|
TF:M04788_1
|
Factor: IRF-1; motif: STTTCACTTTCNNT; match class: 1
|
4.53680151949136e-05
|
17
|
79
|
4
|
0.0506329113924051
|
0.235294117647059
|
|
TF
|
TF:M01884
|
Factor: IRF-7; motif: AAGWGAA
|
6.03470115262041e-05
|
10868
|
117
|
90
|
0.769230769230769
|
0.00828119249171881
|
|
TF
|
TF:M11678
|
Factor: IRF-7; motif: NNGAAAGTGAAANTR
|
0.000130754117367601
|
179
|
90
|
8
|
0.0888888888888889
|
0.0446927374301676
|
|
TF
|
TF:M11383
|
Factor: ESE-1; motif: ACCAGGAAGTANNNNNNNAAWAA
|
0.00017755368396212
|
3886
|
139
|
51
|
0.366906474820144
|
0.0131240349974267
|
|
TF
|
TF:M05989
|
Factor: ZNF275; motif: KGGGMCWTTAGA
|
0.000197286057391471
|
36
|
15
|
3
|
0.2
|
0.0833333333333333
|
|
TF
|
TF:M00062
|
Factor: IRF-1; motif: SAAAAGYGAAACC
|
0.000220775807192332
|
576
|
138
|
16
|
0.115942028985507
|
0.0277777777777778
|
|
TF
|
TF:M00054
|
Factor: NF-kappaB; motif: GGGAMTTYCC
|
0.000222107272891943
|
2444
|
41
|
17
|
0.414634146341463
|
0.00695581014729951
|
|
TF
|
TF:M00747
|
Factor: IRF-1; motif: TTCACTT
|
0.000248032717191808
|
5208
|
117
|
54
|
0.461538461538462
|
0.01036866359447
|
|
TF
|
TF:M07314
|
Factor: Blimp-1; motif: RGRAAGNGAAAG
|
0.000248032717191808
|
1460
|
93
|
21
|
0.225806451612903
|
0.0143835616438356
|
|
TF
|
TF:M03557
|
Factor: P50; motif: GGRRANTCCCNN
|
0.000248496548124828
|
4967
|
63
|
33
|
0.523809523809524
|
0.00664384940608013
|
|
TF
|
TF:M02106
|
Factor: NF-YA; motif: CRGCCAATCAGNRN
|
0.000295899329989728
|
1741
|
17
|
9
|
0.529411764705882
|
0.00516944284893739
|
|
TF
|
TF:M00051
|
Factor: NF-kappaB; motif: GGGGATYCCC
|
0.0003115950134425
|
6413
|
26
|
20
|
0.769230769230769
|
0.00311866521128957
|
|
TF
|
TF:M03557_1
|
Factor: P50; motif: GGRRANTCCCNN; match class: 1
|
0.0003115950134425
|
1292
|
39
|
12
|
0.307692307692308
|
0.00928792569659443
|
|
TF
|
TF:M09668
|
Factor: RelA-p65; motif: GGGRNTTTCCM
|
0.0003115950134425
|
3576
|
110
|
40
|
0.363636363636364
|
0.0111856823266219
|
|
TF
|
TF:M03882
|
Factor: RelB:p50; motif: RGAAANTCCCYNNHGC
|
0.0003115950134425
|
4474
|
55
|
28
|
0.509090909090909
|
0.00625838176128744
|
|
TF
|
TF:M01884_1
|
Factor: IRF-7; motif: AAGWGAA; match class: 1
|
0.000514431825159454
|
4556
|
106
|
45
|
0.424528301886792
|
0.00987708516242318
|
|
TF
|
TF:M00209
|
Factor: NF-Y; motif: NCTGATTGGYTASY
|
0.000526345952514393
|
3776
|
25
|
15
|
0.6
|
0.00397245762711864
|
|
TF
|
TF:M00194_1
|
Factor: NF-kappaB; motif: NGGGGAMTTTCCNN; match class: 1
|
0.000537354555937838
|
385
|
107
|
11
|
0.102803738317757
|
0.0285714285714286
|
|
TF
|
TF:M11679_1
|
Factor: IRF-7; motif: NCGAAANCGAAANYN; match class: 1
|
0.00059674824994574
|
23
|
35
|
3
|
0.0857142857142857
|
0.130434782608696
|
|
TF
|
TF:M00775
|
Factor: NF-Y; motif: NNNNRRCCAATSR
|
0.000658971077082496
|
1428
|
17
|
8
|
0.470588235294118
|
0.00560224089635854
|
|
TF
|
TF:M10080_1
|
Factor: STAT2; motif: RRGRAANNGAAACTGAAAN; match class: 1
|
0.000658971077082496
|
11
|
79
|
3
|
0.0379746835443038
|
0.272727272727273
|
|
TF
|
TF:M02107
|
Factor: NF-YC; motif: NRGCCAATYAGMGC
|
0.00069011037508733
|
1001
|
17
|
7
|
0.411764705882353
|
0.00699300699300699
|
|
TF
|
TF:M00208
|
Factor: NF-kappaB; motif: NGGGACTTTCCA
|
0.000750749846974676
|
3757
|
53
|
24
|
0.452830188679245
|
0.00638807559222784
|
|
TF
|
TF:M03545_1
|
Factor: c-Rel; motif: NGGGAATYTCCN; match class: 1
|
0.000750749846974676
|
1011
|
53
|
12
|
0.226415094339623
|
0.0118694362017804
|
|
TF
|
TF:M04849
|
Factor: RelA-p65; motif: AAASTCCC
|
0.000794176610926652
|
6240
|
44
|
28
|
0.636363636363636
|
0.00448717948717949
|
|
TF
|
TF:M01224
|
Factor: P50:RELA-P65; motif: GGANTTYCCCWN
|
0.000938970104945619
|
2329
|
136
|
34
|
0.25
|
0.0145985401459854
|
|
TF
|
TF:M11673
|
Factor: IRF-5; motif: NCGAAACCGAAACY
|
0.00121259617738235
|
57
|
114
|
5
|
0.043859649122807
|
0.087719298245614
|
|
TF
|
TF:M01066
|
Factor: Blimp-1; motif: NRGRAAGKGAAAGKRN
|
0.00127154699245078
|
411
|
111
|
11
|
0.0990990990990991
|
0.0267639902676399
|
|
TF
|
TF:M01883_1
|
Factor: IRF-4; motif: GAAARTA; match class: 1
|
0.00158833299068059
|
2948
|
94
|
30
|
0.319148936170213
|
0.0101763907734057
|
|
TF
|
TF:M00774_1
|
Factor: NF-kappaB; motif: NGGGANTTYCCMNNNN; match class: 1
|
0.00182264185312236
|
192
|
19
|
4
|
0.210526315789474
|
0.0208333333333333
|
|
TF
|
TF:M04020_1
|
Factor: IRF8; motif: NCGAAACCGAAACT; match class: 1
|
0.00188179681442902
|
45
|
79
|
4
|
0.0506329113924051
|
0.0888888888888889
|
|
TF
|
TF:M04016
|
Factor: IRF5; motif: CCGAAACCGAAACY
|
0.00190193835583645
|
314
|
125
|
10
|
0.08
|
0.0318471337579618
|
|
TF
|
TF:M01223_1
|
Factor: P50:P50; motif: NRGGGAMTNCCCN; match class: 1
|
0.00196029777382715
|
161
|
23
|
4
|
0.173913043478261
|
0.0248447204968944
|
|
TF
|
TF:M00155
|
Factor: ARP-1; motif: TGARCCYTTGAMCCCW
|
0.00196029777382715
|
5581
|
38
|
23
|
0.605263157894737
|
0.00412112524637162
|
|
TF
|
TF:M01665_1
|
Factor: IRF-8; motif: AGTTTCW; match class: 1
|
0.00227605453456789
|
721
|
122
|
15
|
0.122950819672131
|
0.0208044382801664
|
|
TF
|
TF:M08895
|
Factor: PITX; motif: TAATCCCN
|
0.00305002774453606
|
854
|
17
|
6
|
0.352941176470588
|
0.00702576112412178
|
|
TF
|
TF:M00051_1
|
Factor: NF-kappaB; motif: GGGGATYCCC; match class: 1
|
0.00315243311124505
|
2797
|
25
|
12
|
0.48
|
0.00429031104755095
|
|
TF
|
TF:M04054_1
|
Factor: NF-kappaB; motif: AGGGGAWTCCCCT; match class: 1
|
0.00383803163685216
|
1295
|
23
|
8
|
0.347826086956522
|
0.00617760617760618
|
|
TF
|
TF:M11807
|
Factor: RAR-gamma; motif: RAGGTCATGACCTY
|
0.00400487533215996
|
2046
|
132
|
29
|
0.21969696969697
|
0.0141739980449658
|
|
TF
|
TF:M03563
|
Factor: RelA-p65; motif: GGGANTTTCCNN
|
0.00447303334329128
|
4643
|
139
|
53
|
0.381294964028777
|
0.0114150333835882
|
|
TF
|
TF:M00747_1
|
Factor: IRF-1; motif: TTCACTT; match class: 1
|
0.00453410122979223
|
933
|
113
|
16
|
0.141592920353982
|
0.0171489817792069
|
|
TF
|
TF:M10963
|
Factor: RHOXF1; motif: NGGATCAN
|
0.00485097930843662
|
2641
|
66
|
21
|
0.318181818181818
|
0.00795153351003408
|
|
TF
|
TF:M10004
|
Factor: NF-kappaB2; motif: GGGRAANYCCC
|
0.00489818994686276
|
1073
|
110
|
17
|
0.154545454545455
|
0.0158434296365331
|
|
TF
|
TF:M11674
|
Factor: IRF-3; motif: GCNGTTTCCWGGAAACNGAAAC
|
0.00492648447276117
|
4623
|
55
|
26
|
0.472727272727273
|
0.00562405364481938
|
|
TF
|
TF:M11800_1
|
Factor: NR1B2; motif: RAGGTCRTGACCTY; match class: 1
|
0.00495011588554227
|
987
|
23
|
7
|
0.304347826086957
|
0.00709219858156028
|
|
TF
|
TF:M01260_1
|
Factor: STAT1; motif: NNTTTCYNGGAARNNNNNNNNN; match class: 1
|
0.00500705736681396
|
657
|
128
|
14
|
0.109375
|
0.0213089802130898
|
|
TF
|
TF:M03547
|
Factor: ER-alpha; motif: TGACCYN
|
0.00507661188516162
|
4712
|
43
|
22
|
0.511627906976744
|
0.00466893039049236
|
|
TF
|
TF:M07222
|
Factor: NFYA; motif: AGNSYKCTGATTGGTNNR
|
0.00519477161857941
|
714
|
22
|
6
|
0.272727272727273
|
0.00840336134453781
|
|
TF
|
TF:M00621
|
Factor: C/EBPdelta; motif: MATTKCNTMAYY
|
0.0058606406080659
|
3888
|
68
|
27
|
0.397058823529412
|
0.00694444444444444
|
|
TF
|
TF:M09839
|
Factor: ATF4; motif: ATTGCATCAT
|
0.00707744444057751
|
2145
|
4
|
4
|
1
|
0.00186480186480186
|
|
TF
|
TF:M05528
|
Factor: ZAC; motif: KGWTGGGSAT
|
0.00707744444057751
|
147
|
14
|
3
|
0.214285714285714
|
0.0204081632653061
|
|
TF
|
TF:M09652
|
Factor: rfx5; motif: STCNNCWGTTGCYRGGNNRA
|
0.00771308876854271
|
981
|
91
|
14
|
0.153846153846154
|
0.0142711518858308
|
|
TF
|
TF:M11803_1
|
Factor: NR1B2; motif: RAGGTCATGACCTN; match class: 1
|
0.00783601694417448
|
915
|
19
|
6
|
0.315789473684211
|
0.00655737704918033
|
|
TF
|
TF:M10526
|
Factor: Blimp-1; motif: RRGNGAAAGNRANNN
|
0.00790562503969781
|
2327
|
61
|
18
|
0.295081967213115
|
0.00773528147829824
|
|
TF
|
TF:M08359
|
Factor: HOXD12:Elk-1; motif: NRNCGGAAGTCGTAAAN
|
0.00854968123288167
|
6152
|
88
|
44
|
0.5
|
0.00715214564369311
|
|
TF
|
TF:M07045_1
|
Factor: IRF-1; motif: NNRAAANNGAAASN; match class: 1
|
0.00856343646692278
|
19
|
116
|
3
|
0.0258620689655172
|
0.157894736842105
|
|
TF
|
TF:M10526_1
|
Factor: Blimp-1; motif: RRGNGAAAGNRANNN; match class: 1
|
0.00856343646692278
|
286
|
137
|
9
|
0.0656934306569343
|
0.0314685314685315
|
|
TF
|
TF:M07355
|
Factor: GR; motif: AGAACAN
|
0.00856343646692278
|
5862
|
105
|
49
|
0.466666666666667
|
0.00835892186966906
|
|
TF
|
TF:M05548
|
Factor: PLAGL2; motif: AAGGACC
|
0.00911695661663607
|
4194
|
15
|
10
|
0.666666666666667
|
0.00238435860753457
|
|
TF
|
TF:M01653_1
|
Factor: HMGIY; motif: NGWWATTN; match class: 1
|
0.00911695661663607
|
1496
|
35
|
10
|
0.285714285714286
|
0.00668449197860963
|
|
TF
|
TF:M04756_1
|
Factor: sin3A; motif: TGTCCNNGGTGCTG; match class: 1
|
0.00920040204642112
|
7714
|
25
|
19
|
0.76
|
0.00246305418719212
|
|
TF
|
TF:M01728_1
|
Factor: EAR2; motif: YGNNCTTTGNCCTK; match class: 1
|
0.00927253848338919
|
1256
|
10
|
5
|
0.5
|
0.00398089171974522
|
|
TF
|
TF:M07314_1
|
Factor: Blimp-1; motif: RGRAAGNGAAAG; match class: 1
|
0.0100089418404849
|
118
|
91
|
5
|
0.0549450549450549
|
0.0423728813559322
|
|
TF
|
TF:M10046
|
Factor: c-Rel; motif: NRNRGGGRAATKCCA
|
0.0101854360486966
|
4696
|
139
|
52
|
0.37410071942446
|
0.0110732538330494
|
|
TF
|
TF:M00774
|
Factor: NF-kappaB; motif: NGGGANTTYCCMNNNN
|
0.010218672292292
|
1818
|
41
|
12
|
0.292682926829268
|
0.0066006600660066
|
|
TF
|
TF:M06132
|
Factor: ZNF823; motif: NYYACCAGCCCT
|
0.010218672292292
|
55
|
107
|
4
|
0.0373831775700935
|
0.0727272727272727
|
|
TF
|
TF:M09589_1
|
Factor: AR; motif: NGNACANNNTGTTCYNN; match class: 1
|
0.0106720851686209
|
1159
|
11
|
5
|
0.454545454545455
|
0.00431406384814495
|
|
TF
|
TF:M10094
|
Factor: RelA-p65; motif: NWGGGRATTTCCMN
|
0.010851090554504
|
2429
|
41
|
14
|
0.341463414634146
|
0.00576368876080692
|
|
TF
|
TF:M11908_1
|
Factor: POU3F2; motif: TAATKAGNNNNTAATKA; match class: 1
|
0.0111200243446727
|
1278
|
101
|
17
|
0.168316831683168
|
0.013302034428795
|
|
TF
|
TF:M01718
|
Factor: NFATc1; motif: TTTCCWN
|
0.0111654989129912
|
5943
|
14
|
11
|
0.785714285714286
|
0.00185091704526334
|
|
TF
|
TF:M00302
|
Factor: NFAT; motif: NANWGGAAAANN
|
0.0111654989129912
|
805
|
102
|
13
|
0.127450980392157
|
0.0161490683229814
|
|
TF
|
TF:M01151
|
Factor: DMRT7; motif: TTGTTACAWTKTKG
|
0.0111654989129912
|
1009
|
127
|
17
|
0.133858267716535
|
0.0168483647175421
|
|
TF
|
TF:M00240_1
|
Factor: Nkx2-5; motif: TYAAGTG; match class: 1
|
0.0111654989129912
|
877
|
132
|
16
|
0.121212121212121
|
0.0182440136830103
|
|
TF
|
TF:M11853
|
Factor: RORgamma; motif: TAGGTCRTGACCTA
|
0.0113967756525862
|
159
|
16
|
3
|
0.1875
|
0.0188679245283019
|
|
TF
|
TF:M11817_1
|
Factor: T3R-beta; motif: NRRGGTCRTGACCYYN; match class: 1
|
0.0117394695976773
|
1566
|
132
|
23
|
0.174242424242424
|
0.0146871008939974
|
|
TF
|
TF:M01203_1
|
Factor: SPI1; motif: NNAAWGNGGAASTNNNN; match class: 1
|
0.0117634533033348
|
211
|
30
|
4
|
0.133333333333333
|
0.018957345971564
|
|
TF
|
TF:M01197
|
Factor: Elf5; motif: WNAAGGAARTW
|
0.0117634533033348
|
2095
|
139
|
29
|
0.20863309352518
|
0.0138424821002387
|
|
TF
|
TF:M02112
|
Factor: OC-2; motif: TCAATA
|
0.0117842672701313
|
7568
|
35
|
24
|
0.685714285714286
|
0.00317124735729387
|
|
TF
|
TF:M07221
|
Factor: NF-kappaB1; motif: KGGRNTTTCCM
|
0.0117842672701313
|
2417
|
139
|
32
|
0.23021582733813
|
0.0132395531650807
|
|
TF
|
TF:M00053
|
Factor: c-Rel; motif: SGGRNTTTCC
|
0.0120917430654788
|
3672
|
53
|
21
|
0.39622641509434
|
0.00571895424836601
|
|
TF
|
TF:M10003
|
Factor: NF-kappaB1; motif: AGGGAAAK
|
0.0121884559500123
|
1983
|
140
|
28
|
0.2
|
0.0141200201714574
|
|
TF
|
TF:M11807_1
|
Factor: RAR-gamma; motif: RAGGTCATGACCTY; match class: 1
|
0.0122516310135269
|
651
|
19
|
5
|
0.263157894736842
|
0.00768049155145929
|
|
TF
|
TF:M00790_1
|
Factor: HNF-1; motif: WRGTTAATNATTAACNNN; match class: 1
|
0.0122516310135269
|
372
|
49
|
6
|
0.122448979591837
|
0.0161290322580645
|
|
TF
|
TF:M01388
|
Factor: Dlx-5; motif: NNRGYAATTRNYKNNN
|
0.0130544041257247
|
7668
|
106
|
59
|
0.556603773584906
|
0.0076943140323422
|
|
TF
|
TF:M09933_1
|
Factor: FOXP1; motif: TNTGTTTMY; match class: 1
|
0.0130544041257247
|
634
|
115
|
12
|
0.104347826086957
|
0.0189274447949527
|
|
TF
|
TF:M12068
|
Factor: ZNF32; motif: NNGTAACNYGAYACC
|
0.0132626559259519
|
567
|
93
|
10
|
0.10752688172043
|
0.017636684303351
|
|
TF
|
TF:M10046_1
|
Factor: c-Rel; motif: NRNRGGGRAATKCCA; match class: 1
|
0.0138835407357326
|
734
|
48
|
8
|
0.166666666666667
|
0.0108991825613079
|
|
TF
|
TF:M09960
|
Factor: IRF-8; motif: NNAARRGGAASTGAAASTNN
|
0.0138835407357326
|
282
|
66
|
6
|
0.0909090909090909
|
0.0212765957446809
|
|
TF
|
TF:M01665
|
Factor: IRF-8; motif: AGTTTCW
|
0.0145676437169706
|
4771
|
133
|
50
|
0.37593984962406
|
0.0104799832320268
|
|
TF
|
TF:M11378
|
Factor: ESE-1; motif: ACMAGGAAGTANNNNNNNWWAWW
|
0.0150660993609765
|
3049
|
109
|
31
|
0.284403669724771
|
0.0101672679567071
|
|
TF
|
TF:M04354_1
|
Factor: HOXC11; motif: NGYMATAAAAN; match class: 1
|
0.0150660993609765
|
339
|
129
|
9
|
0.0697674418604651
|
0.0265486725663717
|
|
TF
|
TF:M11670_1
|
Factor: IRF-5; motif: NYGAAACCGAAACY; match class: 1
|
0.015182282545975
|
36
|
79
|
3
|
0.0379746835443038
|
0.0833333333333333
|
|
TF
|
TF:M10126
|
Factor: ZNF134; motif: GCCTNNNCTMATCAGKTGAAGG
|
0.0155108237458609
|
223
|
119
|
7
|
0.0588235294117647
|
0.031390134529148
|
|
TF
|
TF:M04054
|
Factor: NF-kappaB; motif: AGGGGAWTCCCCT
|
0.0164263110744189
|
2924
|
25
|
11
|
0.44
|
0.00376196990424077
|
|
TF
|
TF:M10848
|
Factor: HOXB13; motif: NCCAATAAAAC
|
0.018002743654089
|
102
|
30
|
3
|
0.1
|
0.0294117647058824
|
|
TF
|
TF:M10841
|
Factor: HOXC13; motif: NCCAATAAAAC
|
0.018002743654089
|
102
|
30
|
3
|
0.1
|
0.0294117647058824
|
|
TF
|
TF:M10079_1
|
Factor: STAT1; motif: NCCNTTTCCNGGAAANCNC; match class: 1
|
0.0190120024828658
|
1164
|
128
|
18
|
0.140625
|
0.0154639175257732
|
|
TF
|
TF:M07323_1
|
Factor: IRF-4; motif: KRAAMNGAAANYN; match class: 1
|
0.0191055704356127
|
39
|
80
|
3
|
0.0375
|
0.0769230769230769
|
|
TF
|
TF:M11741_1
|
Factor: COUP-TF1; motif: RRGGTCRNTGACCYY; match class: 1
|
0.0191517784657492
|
1014
|
29
|
7
|
0.241379310344828
|
0.00690335305719921
|
|
TF
|
TF:M11777
|
Factor: RXR-gamma; motif: NRGGTCAYGACCYN
|
0.0191517784657492
|
138
|
23
|
3
|
0.130434782608696
|
0.0217391304347826
|
|
TF
|
TF:M08774
|
Factor: HOXD4; motif: NNYMATTANN
|
0.0192565606002649
|
4001
|
90
|
32
|
0.355555555555556
|
0.00799800049987503
|
|
TF
|
TF:M06142
|
Factor: ZNF708; motif: GGAAAGTARA
|
0.0202598309838098
|
164
|
46
|
4
|
0.0869565217391304
|
0.024390243902439
|
|
TF
|
TF:M00054_1
|
Factor: NF-kappaB; motif: GGGAMTTYCC; match class: 1
|
0.0205413430727011
|
712
|
53
|
8
|
0.150943396226415
|
0.0112359550561798
|
|
TF
|
TF:M01356
|
Factor: PMX2B; motif: NNNAATTAATTAANNNG
|
0.0206416073142583
|
3215
|
106
|
31
|
0.292452830188679
|
0.00964230171073095
|
|
TF
|
TF:M12334
|
Factor: ZBTB20; motif: NNAAYGTATRKN
|
0.0207723279446488
|
1214
|
94
|
15
|
0.159574468085106
|
0.0123558484349259
|
|
TF
|
TF:M11783
|
Factor: RXR-beta; motif: RRGGTCATGACCYY
|
0.0209912238616643
|
144
|
23
|
3
|
0.130434782608696
|
0.0208333333333333
|
|
TF
|
TF:M09899
|
Factor: ehf; motif: NNANSAGGAAGTNNN
|
0.0213720404359395
|
457
|
82
|
8
|
0.0975609756097561
|
0.0175054704595186
|
|
TF
|
TF:M08359_1
|
Factor: HOXD12:Elk-1; motif: NRNCGGAAGTCGTAAAN; match class: 1
|
0.0213720404359395
|
1166
|
88
|
14
|
0.159090909090909
|
0.0120068610634648
|
|
TF
|
TF:M09629_1
|
Factor: IRF-1; motif: NAAANNGAAAGTGAAASTRN; match class: 1
|
0.0219607253629208
|
29
|
116
|
3
|
0.0258620689655172
|
0.103448275862069
|
|
TF
|
TF:M04017
|
Factor: IRF5; motif: NACCGAAACYA
|
0.0219607253629208
|
195
|
70
|
5
|
0.0714285714285714
|
0.0256410256410256
|
|
TF
|
TF:M04013
|
Factor: HSFY1; motif: TTCGAANSRTTCGAA
|
0.022065589825898
|
962
|
23
|
6
|
0.260869565217391
|
0.00623700623700624
|
|
TF
|
TF:M08302_1
|
Factor: FOXO1A:Net; motif: RWMAACAGGAAGTN; match class: 1
|
0.0223046386287566
|
2779
|
53
|
17
|
0.320754716981132
|
0.0061173083843109
|
|
TF
|
TF:M09663_1
|
Factor: STAT1; motif: YCMTTTCCNGGAAAWCNCATNN; match class: 1
|
0.0223046386287566
|
554
|
53
|
7
|
0.132075471698113
|
0.0126353790613718
|
|
TF
|
TF:M04279
|
Factor: ALX3; motif: NNYAATTANN
|
0.0225789804189304
|
3934
|
100
|
34
|
0.34
|
0.00864260294865277
|
|
TF
|
TF:M04055
|
Factor: NFKB2; motif: NGGGGAWTCCCCN
|
0.0227074029888114
|
55
|
62
|
3
|
0.0483870967741935
|
0.0545454545454545
|
|
TF
|
TF:M04055_1
|
Factor: NFKB2; motif: NGGGGAWTCCCCN; match class: 1
|
0.0227074029888114
|
55
|
62
|
3
|
0.0483870967741935
|
0.0545454545454545
|
|
TF
|
TF:M04731
|
Factor: Blimp-1; motif: NACTTTCAC
|
0.0237328458602237
|
3997
|
114
|
38
|
0.333333333333333
|
0.00950713034776082
|
|
TF
|
TF:M12207
|
Factor: GATA-4; motif: NNGATAASNN
|
0.0237328458602237
|
2844
|
95
|
26
|
0.273684210526316
|
0.00914205344585091
|
|
TF
|
TF:M01379
|
Factor: HNF-1alpha; motif: NNKNNGTTAACTAANNN
|
0.0241489458185346
|
3368
|
6
|
5
|
0.833333333333333
|
0.00148456057007126
|
|
TF
|
TF:M08957_1
|
Factor: NURR1:RXR-ALPHA; motif: NRGGTCRTTGACCYN; match class: 1
|
0.0246179345379357
|
3226
|
39
|
15
|
0.384615384615385
|
0.004649721016739
|
|
TF
|
TF:M04450_1
|
Factor: VSX1; motif: NGCYAATTRNN; match class: 1
|
0.0249660867724284
|
3267
|
106
|
31
|
0.292452830188679
|
0.00948882767064585
|
|
TF
|
TF:M02116_1
|
Factor: Sox-10; motif: NACAAWG; match class: 1
|
0.0249753837766241
|
8039
|
94
|
54
|
0.574468085106383
|
0.00671725338972509
|
|
TF
|
TF:M11974
|
Factor: rfx5; motif: NYRGCAACSGTTGCYAN
|
0.025210069798962
|
629
|
14
|
4
|
0.285714285714286
|
0.00635930047694754
|
|
TF
|
TF:M02117
|
Factor: STAT4; motif: TTMNNRGAAA
|
0.025210069798962
|
1353
|
53
|
11
|
0.207547169811321
|
0.00813008130081301
|
|
TF
|
TF:M10843
|
Factor: HOXD13; motif: NSYAATAAAAN
|
0.0255437213656835
|
968
|
73
|
11
|
0.150684931506849
|
0.0113636363636364
|
|
TF
|
TF:M09813_1
|
Factor: MYB; motif: NNCAACTGNN; match class: 1
|
0.0255437213656835
|
979
|
32
|
7
|
0.21875
|
0.00715015321756895
|
|
TF
|
TF:M00746
|
Factor: Elf-1; motif: RNWMBAGGAART
|
0.0255437213656835
|
3365
|
27
|
12
|
0.444444444444444
|
0.00356612184249629
|
|
TF
|
TF:M11676_1
|
Factor: IRF-3; motif: NGGAAANGGAAASNGAAACN; match class: 1
|
0.0258096391535786
|
41
|
89
|
3
|
0.0337078651685393
|
0.0731707317073171
|
|
TF
|
TF:M00638
|
Factor: HNF4alpha; motif: VTGAACTTTGMMB
|
0.0260393734932712
|
2814
|
58
|
18
|
0.310344827586207
|
0.00639658848614072
|
|
TF
|
TF:M09903
|
Factor: elf5; motif: NNNANSAGGAAGTNN
|
0.0260393734932712
|
1243
|
41
|
9
|
0.219512195121951
|
0.00724054706355591
|
|
TF
|
TF:M10832
|
Factor: HOXA13; motif: NCCAATAAAAN
|
0.0266256537199694
|
679
|
73
|
9
|
0.123287671232877
|
0.0132547864506627
|
|
TF
|
TF:M09589
|
Factor: AR; motif: NGNACANNNTGTTCYNN
|
0.0267470236077955
|
5130
|
17
|
11
|
0.647058823529412
|
0.00214424951267057
|
|
TF
|
TF:M00412
|
Factor: AREB6; motif: NNYNYACCTGWVT
|
0.0267470236077955
|
4738
|
105
|
40
|
0.380952380952381
|
0.00844238075137189
|
|
TF
|
TF:M07204
|
Factor: AR; motif: ARGAACANNNTGTNC
|
0.0267470236077955
|
4906
|
47
|
22
|
0.468085106382979
|
0.00448430493273543
|
|
TF
|
TF:M04903
|
Factor: HNF-4alpha; motif: AGTCCAAR
|
0.0267470236077955
|
6987
|
23
|
16
|
0.695652173913043
|
0.0022899670817232
|
|
TF
|
TF:M11765_1
|
Factor: TR2; motif: NRGGTCRYGACCYN; match class: 1
|
0.0268913574359263
|
1431
|
23
|
7
|
0.304347826086957
|
0.00489168413696716
|
|
TF
|
TF:M05946_1
|
Factor: PRDM16; motif: KGGTCATRACCM; match class: 1
|
0.0275852409977403
|
2282
|
24
|
9
|
0.375
|
0.00394390885188431
|
|
TF
|
TF:M07229_1
|
Factor: STAT3; motif: NTTCYKGGAAN; match class: 1
|
0.0277361285690379
|
969
|
122
|
15
|
0.122950819672131
|
0.0154798761609907
|
|
TF
|
TF:M00158
|
Factor: COUP-TF,; motif: TGAMCTTTGMMCYT
|
0.0281915974310228
|
2215
|
116
|
25
|
0.21551724137931
|
0.0112866817155756
|
|
TF
|
TF:M04498_1
|
Factor: VDR; motif: GRGTTCATYGRGTTCA; match class: 1
|
0.0286088096861393
|
876
|
11
|
4
|
0.363636363636364
|
0.0045662100456621
|
|
TF
|
TF:M01362
|
Factor: CART1; motif: NGNNYTAATTARTNNNN
|
0.029089544034154
|
4558
|
106
|
39
|
0.367924528301887
|
0.00855638437911365
|
|
TF
|
TF:M08817_1
|
Factor: HOXD13; motif: WNNAATAAWAY; match class: 1
|
0.0292188643082282
|
1646
|
105
|
19
|
0.180952380952381
|
0.011543134872418
|
|
TF
|
TF:M04327_1
|
Factor: GSX1; motif: NNYMATTANN; match class: 1
|
0.0293458470298593
|
5540
|
106
|
45
|
0.424528301886792
|
0.00812274368231047
|
|
TF
|
TF:M07066
|
Factor: STAT6; motif: TTCYYNGGAAN
|
0.0296325038007278
|
1849
|
35
|
10
|
0.285714285714286
|
0.00540832882639264
|
|
TF
|
TF:M04611_1
|
Factor: FOXM1; motif: NAGASTGATTA; match class: 1
|
0.0296325038007278
|
3507
|
131
|
38
|
0.290076335877863
|
0.0108354719133162
|
|
TF
|
TF:M11895_1
|
Factor: Oct-2; motif: NTATGCGCATAN; match class: 1
|
0.0297772748903679
|
1376
|
88
|
15
|
0.170454545454545
|
0.0109011627906977
|
|
TF
|
TF:M00459_1
|
Factor: STAT5B; motif: NAWTTCYNGGAAWTN; match class: 1
|
0.0299699072277418
|
1896
|
128
|
24
|
0.1875
|
0.0126582278481013
|
|
TF
|
TF:M12067
|
Factor: ZNF32; motif: NYGTAACNYGAYACC
|
0.0300184801416989
|
164
|
93
|
5
|
0.0537634408602151
|
0.0304878048780488
|
|
TF
|
TF:M04249_1
|
Factor: FOXJ3; motif: RTAAACAA; match class: 1
|
0.0303595377670342
|
603
|
138
|
12
|
0.0869565217391304
|
0.0199004975124378
|
|
TF
|
TF:M01043_1
|
Factor: CSX; motif: TSYCACTTSM; match class: 1
|
0.0303595377670342
|
1199
|
71
|
12
|
0.169014084507042
|
0.0100083402835696
|
|
TF
|
TF:M07308
|
Factor: Sox-2; motif: NNNANAACAAWGRNN
|
0.0320175865125575
|
2054
|
67
|
16
|
0.238805970149254
|
0.00778967867575463
|
|
TF
|
TF:M04468
|
Factor: HNF4A; motif: RRGTCCAAAGGTCAN
|
0.032145395889171
|
1186
|
113
|
16
|
0.141592920353982
|
0.0134907251264755
|
|
TF
|
TF:M00070
|
Factor: Tal-1beta:ITF-2; motif: NNNAACAGATGKTNNN
|
0.032145395889171
|
1763
|
92
|
18
|
0.195652173913043
|
0.0102098695405559
|
|
TF
|
TF:M00052
|
Factor: NF-kappaB; motif: GGGRATTTCC
|
0.0323246796869833
|
1874
|
41
|
11
|
0.268292682926829
|
0.00586979722518677
|
|
TF
|
TF:M01000
|
Factor: AIRE; motif: GGTTATTAATTGGTTATATTGGTTD
|
0.0323454157904544
|
309
|
133
|
8
|
0.0601503759398496
|
0.0258899676375405
|
|
TF
|
TF:M10058_1
|
Factor: Six-2; motif: KGAAAYNTGAKMC; match class: 1
|
0.0323644079349178
|
75
|
122
|
4
|
0.0327868852459016
|
0.0533333333333333
|
|
TF
|
TF:M00974
|
Factor: SMAD; motif: TNGNCAGACWN
|
0.0324564908802036
|
4535
|
64
|
26
|
0.40625
|
0.00573318632855568
|
|
TF
|
TF:M02271
|
Factor: HOXA5; motif: CDBWAATK
|
0.0324564908802036
|
1928
|
46
|
12
|
0.260869565217391
|
0.00622406639004149
|
|
TF
|
TF:M07988
|
Factor: EMX1; motif: NNTAATKANN
|
0.0330969974587895
|
3512
|
106
|
32
|
0.30188679245283
|
0.00911161731207289
|
|
TF
|
TF:M02086_1
|
Factor: Cdx-1; motif: TTTATK; match class: 1
|
0.0336533676151169
|
9742
|
95
|
62
|
0.652631578947368
|
0.0063641962636009
|
|
TF
|
TF:M00194
|
Factor: NF-kappaB; motif: NGGGGAMTTTCCNN
|
0.0337716891311758
|
2412
|
53
|
15
|
0.283018867924528
|
0.00621890547263682
|
|
TF
|
TF:M01223
|
Factor: P50:P50; motif: NRGGGAMTNCCCN
|
0.0337716891311758
|
1283
|
136
|
19
|
0.139705882352941
|
0.014809041309431
|
|
TF
|
TF:M01388_1
|
Factor: Dlx-5; motif: NNRGYAATTRNYKNNN; match class: 1
|
0.0337716891311758
|
5764
|
106
|
46
|
0.433962264150943
|
0.00798056904927134
|
|
TF
|
TF:M04197
|
Factor: ATF4; motif: NNATGAYGCAATM
|
0.0341119302728259
|
2164
|
3
|
3
|
1
|
0.00138632162661738
|
|
TF
|
TF:M07614
|
Factor: Sox-4; motif: ACAAWGNRNYN
|
0.0342234823660491
|
1860
|
48
|
12
|
0.25
|
0.00645161290322581
|
|
TF
|
TF:M05467
|
Factor: Sall1; motif: NGGTCCKRGKRA
|
0.0369704338721174
|
4211
|
52
|
21
|
0.403846153846154
|
0.00498693896936595
|
|
TF
|
TF:M01308_1
|
Factor: Sox-4; motif: AACAAWGR; match class: 1
|
0.037110015839733
|
107
|
90
|
4
|
0.0444444444444444
|
0.0373831775700935
|
|
TF
|
TF:M01124_1
|
Factor: Oct-4; motif: ATTGWSWTGCWAAWN; match class: 1
|
0.037110015839733
|
1757
|
94
|
18
|
0.191489361702128
|
0.0102447353443369
|
|
TF
|
TF:M02016
|
Factor: HNF-4alpha; motif: GNNCAAAGKYCANNN
|
0.0371603312326913
|
2384
|
20
|
8
|
0.4
|
0.00335570469798658
|
|
TF
|
TF:M11674_1
|
Factor: IRF-3; motif: GCNGTTTCCWGGAAACNGAAAC; match class: 1
|
0.0371603312326913
|
1663
|
140
|
23
|
0.164285714285714
|
0.0138304269392664
|
|
TF
|
TF:M01010_1
|
Factor: HMGIY; motif: NNKKNAWTTTNYTNN; match class: 1
|
0.0385040862280182
|
3147
|
86
|
25
|
0.290697674418605
|
0.00794407372100413
|
|
TF
|
TF:M04306
|
Factor: DPRX; motif: NNGGATTANN
|
0.0385040862280182
|
4886
|
95
|
37
|
0.389473684210526
|
0.00757265656979124
|
|
TF
|
TF:M10637
|
Factor: Nkx2-3; motif: NSCACTTNNC
|
0.0385040862280182
|
1906
|
67
|
15
|
0.223880597014925
|
0.00786988457502623
|
|
TF
|
TF:M04319
|
Factor: ESX1; motif: NNYAATTANN
|
0.0389715111248335
|
3081
|
102
|
28
|
0.274509803921569
|
0.00908795845504706
|
|
TF
|
TF:M11015
|
Factor: ISL2; motif: SCACTTAN
|
0.0389715111248335
|
4207
|
36
|
16
|
0.444444444444444
|
0.00380318516757785
|
|
TF
|
TF:M10687
|
Factor: HOXB7; motif: NGTAATTANN
|
0.0395787434428322
|
942
|
106
|
13
|
0.122641509433962
|
0.0138004246284501
|
|
TF
|
TF:M01003_1
|
Factor: Helios; motif: NNTWGGGANNN; match class: 1
|
0.0395787434428322
|
420
|
103
|
8
|
0.0776699029126214
|
0.019047619047619
|
|
TF
|
TF:M00083
|
Factor: MZF-1; motif: NGNGGGGA
|
0.0401871080506536
|
2867
|
125
|
31
|
0.248
|
0.0108126961981165
|
|
TF
|
TF:M00346
|
Factor: GATA-1; motif: NCWGATAACA
|
0.0401871080506536
|
4376
|
109
|
38
|
0.348623853211009
|
0.00868372943327239
|
|
TF
|
TF:M07432
|
Factor: Sox-2; motif: NNNNNAACAAWGN
|
0.0401871080506536
|
1541
|
134
|
21
|
0.156716417910448
|
0.0136275146009085
|
|
TF
|
TF:M04299
|
Factor: Dlx-1; motif: NNTAATTANN
|
0.0405235248771398
|
2524
|
47
|
14
|
0.297872340425532
|
0.00554675118858954
|
|
TF
|
TF:M07044_1
|
Factor: HNF-1alpha; motif: NGNNAAWNATTAACNN; match class: 1
|
0.0409564335906994
|
147
|
115
|
5
|
0.0434782608695652
|
0.0340136054421769
|
|
TF
|
TF:M00432
|
Factor: TTF1; motif: ASTCAAGTRK
|
0.0414160728066292
|
5394
|
132
|
52
|
0.393939393939394
|
0.0096403411197627
|
|
TF
|
TF:M00622
|
Factor: C/EBPgamma; motif: YTBATTTCARAAW
|
0.041850891938346
|
6887
|
130
|
62
|
0.476923076923077
|
0.00900246841875998
|
|
TF
|
TF:M01733
|
Factor: MZF-1; motif: TGGGGAR
|
0.0429073100315658
|
11391
|
24
|
21
|
0.875
|
0.00184356070582038
|
|
TF
|
TF:M08899_1
|
Factor: SOX; motif: NNNNCWTTGTT; match class: 1
|
0.0441530716473
|
154
|
67
|
4
|
0.0597014925373134
|
0.025974025974026
|
|
TF
|
TF:M09959_1
|
Factor: IRF-4; motif: NAANGRGGAASTGAAASN; match class: 1
|
0.0441530716473
|
395
|
138
|
9
|
0.0652173913043478
|
0.0227848101265823
|
|
TF
|
TF:M02013_1
|
Factor: HNF-1alpha; motif: RGTTAATNWTT; match class: 1
|
0.0443712120991379
|
90
|
115
|
4
|
0.0347826086956522
|
0.0444444444444444
|
|
TF
|
TF:M09630
|
Factor: IRF-4; motif: RRRGRGGAASTGARAS
|
0.0443712120991379
|
954
|
28
|
6
|
0.214285714285714
|
0.00628930817610063
|
|
TF
|
TF:M11800
|
Factor: NR1B2; motif: RAGGTCRTGACCTY
|
0.0443712120991379
|
3716
|
132
|
39
|
0.295454545454545
|
0.0104951560818084
|
|
TF
|
TF:M02096
|
Factor: ipf1; motif: YTAATGN
|
0.0443770522944996
|
2033
|
46
|
12
|
0.260869565217391
|
0.0059026069847516
|
|
TF
|
TF:M00957_1
|
Factor: PR; motif: NNNNNNRGNACNNKNTGTTCTNNNNNN; match class: 1
|
0.0443770522944996
|
559
|
80
|
8
|
0.1
|
0.0143112701252236
|
|
TF
|
TF:M00634_1
|
Factor: GCM; motif: CNNRCCCGCATD; match class: 1
|
0.0447203261069871
|
145
|
72
|
4
|
0.0555555555555556
|
0.0275862068965517
|
|
TF
|
TF:M03563_1
|
Factor: RelA-p65; motif: GGGANTTTCCNN; match class: 1
|
0.0447203261069871
|
876
|
41
|
7
|
0.170731707317073
|
0.00799086757990868
|
|
TF
|
TF:M01808_1
|
Factor: NMYC; motif: CAYCTG; match class: 1
|
0.0447203261069871
|
6149
|
73
|
35
|
0.479452054794521
|
0.00569198243616848
|
|
TF
|
TF:M01043
|
Factor: CSX; motif: TSYCACTTSM
|
0.0447203261069871
|
6282
|
33
|
19
|
0.575757575757576
|
0.00302451448583254
|
|
TF
|
TF:M11267
|
Factor: c-Jun; motif: NATGACKCATN
|
0.0447203261069871
|
3183
|
124
|
33
|
0.266129032258065
|
0.0103675777568332
|
|
TF
|
TF:M09649
|
Factor: Blimp-1; motif: RAAAGTGAAAGTNN
|
0.0447203261069871
|
300
|
116
|
7
|
0.0603448275862069
|
0.0233333333333333
|
|
TF
|
TF:M07338
|
Factor: Sox-10; motif: ACAAWGNN
|
0.0447203261069871
|
1695
|
92
|
17
|
0.184782608695652
|
0.0100294985250737
|
|
TF
|
TF:M04327
|
Factor: GSX1; motif: NNYMATTANN
|
0.0448291115975532
|
5709
|
106
|
45
|
0.424528301886792
|
0.00788229111928534
|
|
TF
|
TF:M10535_1
|
Factor: znf136; motif: CAAGAATWCTATAYCCAG; match class: 1
|
0.0457401908493568
|
2585
|
111
|
26
|
0.234234234234234
|
0.0100580270793037
|
|
TF
|
TF:M09784
|
Factor: RELB; motif: NNATTCCCCNN
|
0.0463408951031356
|
5356
|
140
|
54
|
0.385714285714286
|
0.0100821508588499
|
|
TF
|
TF:M04197_1
|
Factor: ATF4; motif: NNATGAYGCAATM; match class: 1
|
0.0463408951031356
|
197
|
90
|
5
|
0.0555555555555556
|
0.0253807106598985
|
|
TF
|
TF:M11988
|
Factor: NFATc3; motif: NTTTCCRYGGAAAN
|
0.0463907073931982
|
7175
|
53
|
30
|
0.566037735849057
|
0.00418118466898955
|
|
TF
|
TF:M09663
|
Factor: STAT1; motif: YCMTTTCCNGGAAAWCNCATNN
|
0.046771218763942
|
2762
|
137
|
32
|
0.233576642335766
|
0.0115858073859522
|
|
TF
|
TF:M04349
|
Factor: HOXB5; motif: NNYMATTANN
|
0.046771218763942
|
5728
|
106
|
45
|
0.424528301886792
|
0.00785614525139665
|
|
TF
|
TF:M11012
|
Factor: islet1; motif: SCAMTTAN
|
0.046771218763942
|
6170
|
36
|
20
|
0.555555555555556
|
0.00324149108589951
|
|
TF
|
TF:M01393_1
|
Factor: Msx-2; motif: NNNGACYAATTAGYNNT; match class: 1
|
0.046771218763942
|
3967
|
86
|
29
|
0.337209302325581
|
0.00731031005797832
|
|
TF
|
TF:M08199_1
|
Factor: ATF-4:TEF; motif: GGMTGATGCAATM; match class: 1
|
0.046771218763942
|
119
|
90
|
4
|
0.0444444444444444
|
0.0336134453781513
|
|
TF
|
TF:M11816
|
Factor: T3R-beta; motif: NRGGTCAAAGGTCAN
|
0.0471925565501656
|
11742
|
46
|
37
|
0.804347826086957
|
0.00315108158746381
|
|
TF
|
TF:M03852_1
|
Factor: SREBP-2; motif: NTCACCYNNNN; match class: 1
|
0.0471925565501656
|
2246
|
27
|
9
|
0.333333333333333
|
0.00400712377560107
|
|
TF
|
TF:M03849_1
|
Factor: Sox-4; motif: AACAAA; match class: 1
|
0.0475368805721929
|
9090
|
42
|
29
|
0.69047619047619
|
0.00319031903190319
|
|
TF
|
TF:M07417
|
Factor: Elf-1; motif: NNCAGGAAGNNN
|
0.0475368805721929
|
1774
|
67
|
14
|
0.208955223880597
|
0.00789177001127396
|
|
TF
|
TF:M07429_1
|
Factor: SMAD3; motif: CAGACAS; match class: 1
|
0.0482105034769509
|
1381
|
9
|
4
|
0.444444444444444
|
0.00289645184648805
|
|
TF
|
TF:M10082
|
Factor: STAT4; motif: YTTCYYNGMAN
|
0.0484262557755798
|
882
|
53
|
8
|
0.150943396226415
|
0.0090702947845805
|
|
TF
|
TF:M11383_1
|
Factor: ESE-1; motif: ACCAGGAAGTANNNNNNNAAWAA; match class: 1
|
0.0484262557755798
|
589
|
96
|
9
|
0.09375
|
0.0152801358234295
|
|
TF
|
TF:M10042_1
|
Factor: PR; motif: RGNACATTYTGTNCTN; match class: 1
|
0.0484262557755798
|
998
|
6
|
3
|
0.5
|
0.0030060120240481
|
|
TF
|
TF:M00185
|
Factor: NF-Y; motif: TRRCCAATSRN
|
0.0486994014882776
|
870
|
22
|
5
|
0.227272727272727
|
0.00574712643678161
|
|
TF
|
TF:M01107_1
|
Factor: RUSH-1alpha; motif: NNMCWTNKNN; match class: 1
|
0.0489726533254626
|
385
|
119
|
8
|
0.0672268907563025
|
0.0207792207792208
|
|
TF
|
TF:M11638_1
|
Factor: Sox-4; motif: NAACAAAGRN; match class: 1
|
0.049087592856837
|
296
|
90
|
6
|
0.0666666666666667
|
0.0202702702702703
|
|
TF
|
TF:M11317
|
Factor: C/EBPbeta; motif: NRTTGCGYAAYN
|
0.0496163994758783
|
4365
|
4
|
4
|
1
|
0.000916380297823597
|
|
TF
|
TF:M07228_1
|
Factor: STAT1; motif: NTTCYRGGAAA; match class: 1
|
0.0496163994758783
|
99
|
53
|
3
|
0.0566037735849057
|
0.0303030303030303
|
|
TF
|
TF:M09933
|
Factor: FOXP1; motif: TNTGTTTMY
|
0.0499554783794588
|
4164
|
72
|
26
|
0.361111111111111
|
0.00624399615754083
|
|
TF
|
TF:M08190
|
Factor: AR; motif: GGAACGGWACATGTTCT
|
0.0499582452403379
|
7865
|
93
|
51
|
0.548387096774194
|
0.00648442466624285
|
|
TF
|
TF:M12156_1
|
Factor: Sp2; motif: NTAAGYCCCGCCCMCTN; match class: 1
|
0.0499582452403379
|
1002
|
28
|
6
|
0.214285714285714
|
0.00598802395209581
|
|
WP
|
WP:WP2328
|
Allograft Rejection
|
1.15593869382335e-19
|
88
|
31
|
15
|
0.483870967741935
|
0.170454545454545
|
|
WP
|
WP:WP4217
|
Ebola Virus Pathway on Host
|
3.35572703742993e-19
|
130
|
25
|
15
|
0.6
|
0.115384615384615
|
|
WP
|
WP:WP183
|
Proteasome Degradation
|
5.0081709591421e-12
|
64
|
23
|
9
|
0.391304347826087
|
0.140625
|
|
WP
|
WP:WP619
|
Type II interferon signaling (IFNG)
|
1.47971580064131e-11
|
37
|
66
|
10
|
0.151515151515152
|
0.27027027027027
|
|
WP
|
WP:WP4197
|
The human immune response to tuberculosis
|
3.0749759114202e-06
|
23
|
79
|
6
|
0.0759493670886076
|
0.260869565217391
|
|
WP
|
WP:WP5039
|
SARS-CoV-2 Innate Immunity Evasion and Cell-specific immune response
|
0.000346206874071133
|
68
|
123
|
8
|
0.0650406504065041
|
0.117647058823529
|
|
WP
|
WP:WP4877
|
Host-pathogen interaction of human corona viruses - MAPK signaling
|
0.00490659944626974
|
36
|
27
|
3
|
0.111111111111111
|
0.0833333333333333
|
|
WP
|
WP:WP558
|
Complement and Coagulation Cascades
|
0.0209401345490946
|
59
|
109
|
5
|
0.0458715596330275
|
0.0847457627118644
|
|
WP
|
WP:WP75
|
Toll-like Receptor Signaling Pathway
|
0.0209401345490946
|
104
|
123
|
7
|
0.0569105691056911
|
0.0673076923076923
|
|
WP
|
WP:WP2806
|
Human Complement System
|
0.0346330772550996
|
99
|
109
|
6
|
0.055045871559633
|
0.0606060606060606
|
|
WP
|
WP:WP545
|
Complement Activation
|
0.0403852443230244
|
22
|
109
|
3
|
0.0275229357798165
|
0.136363636363636
|