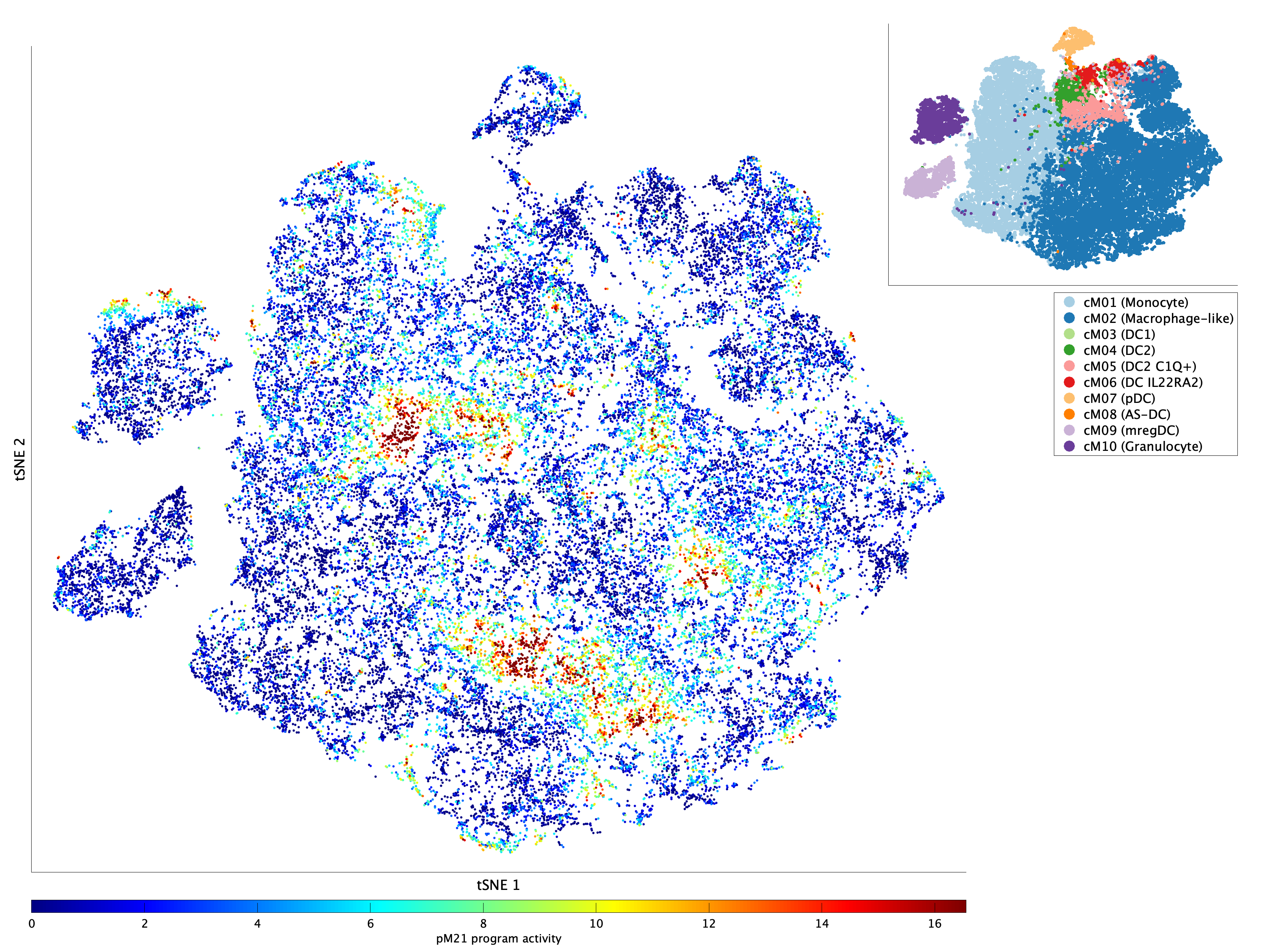
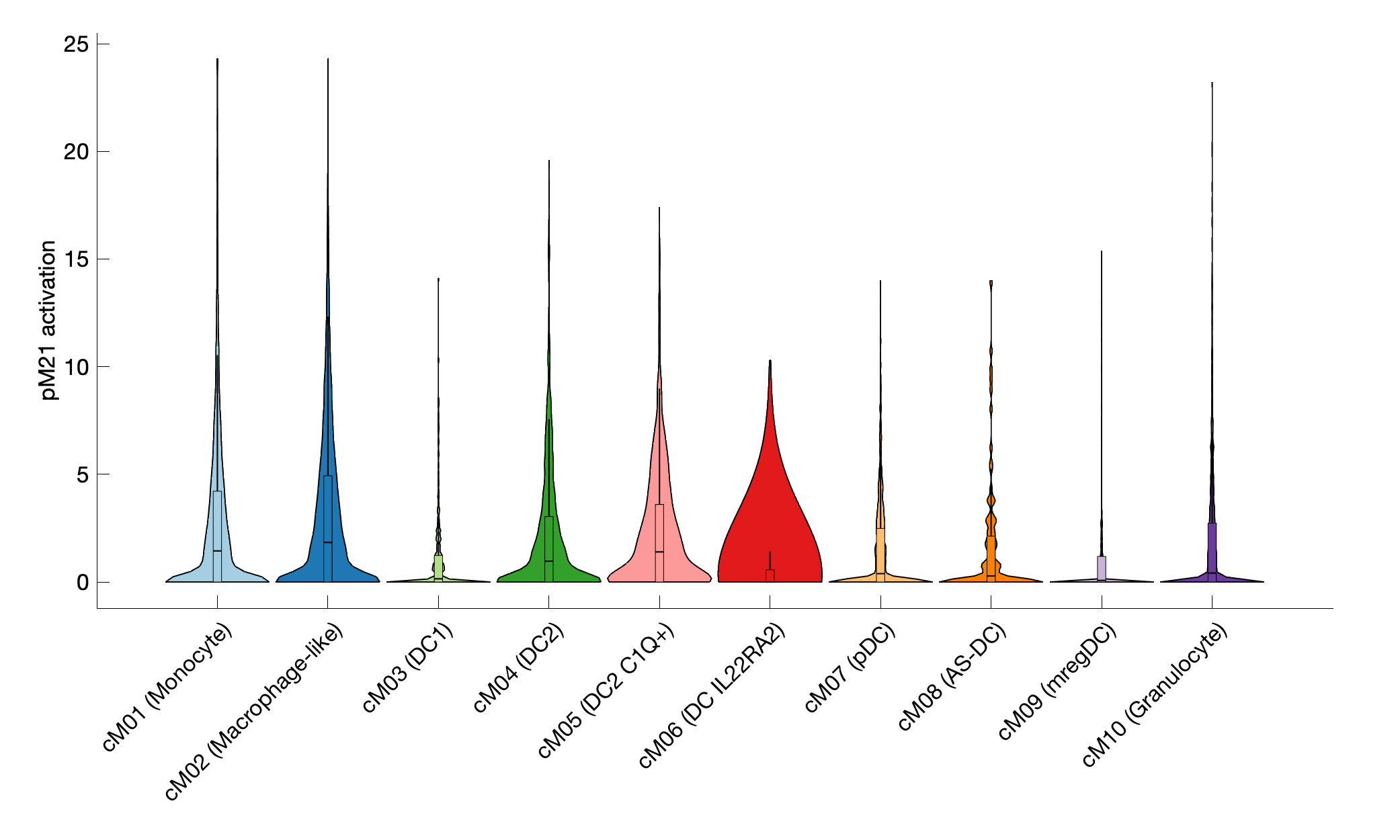
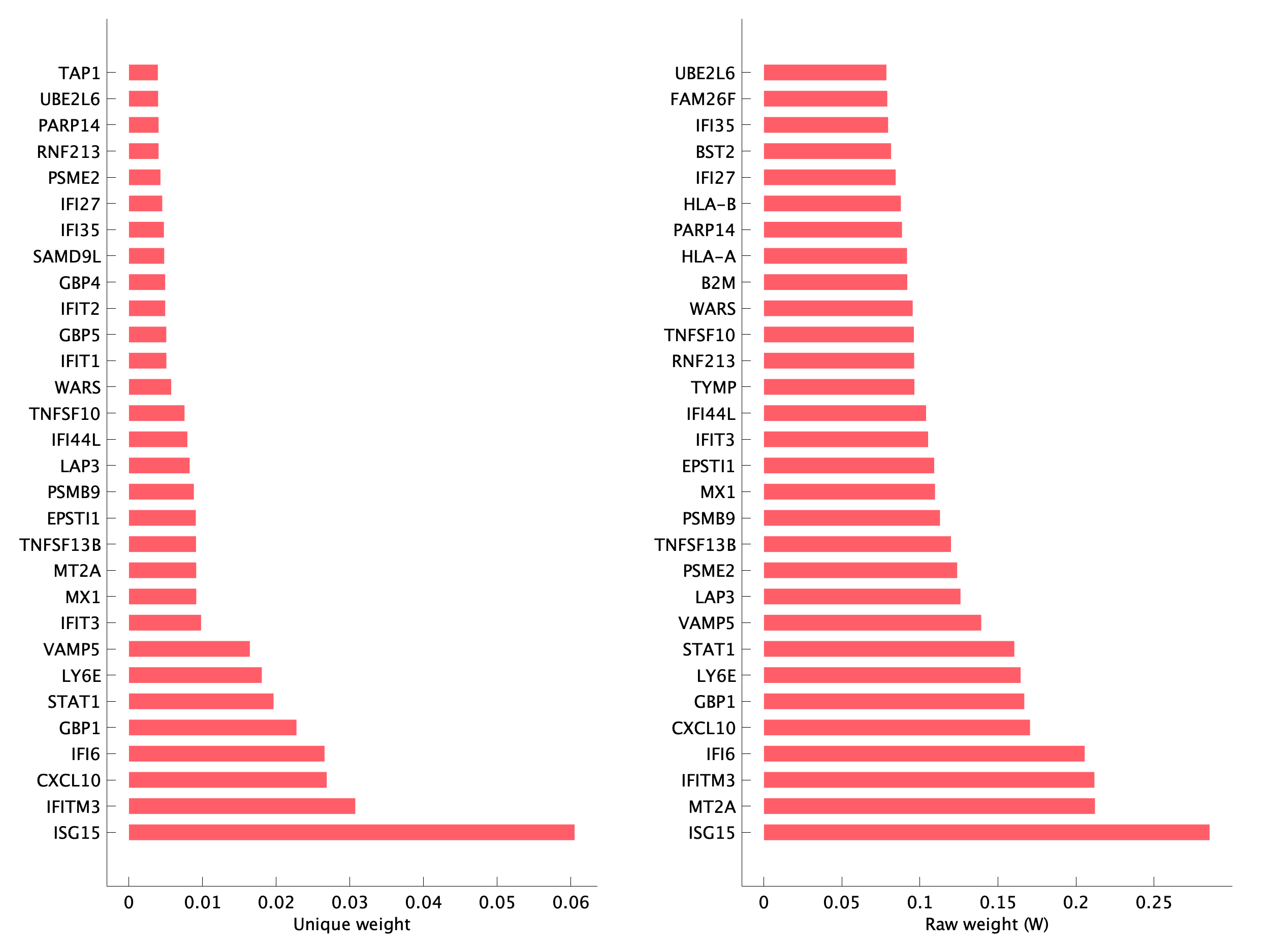
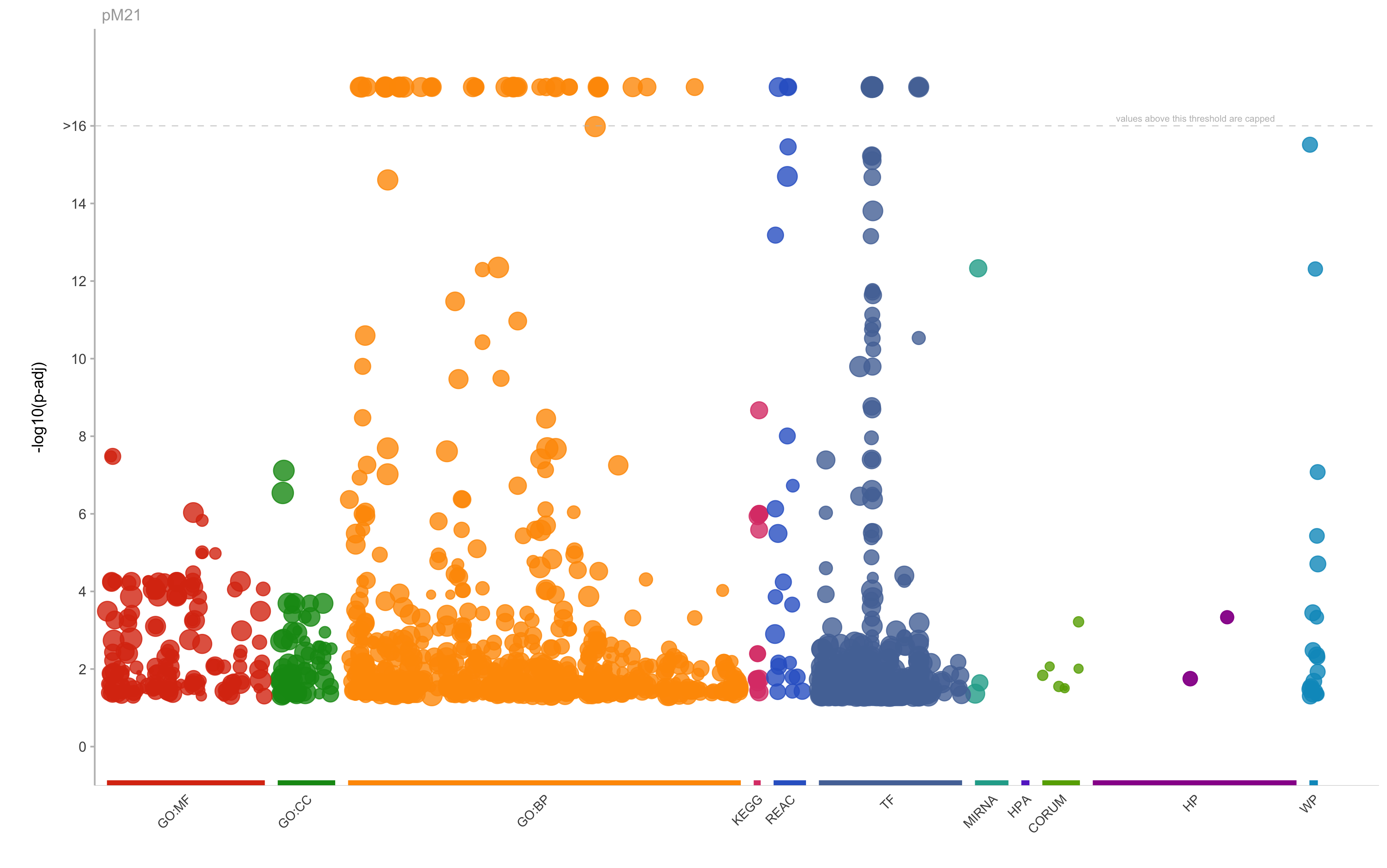
|
CORUM
|
CORUM:7385
|
DTX3L-PARP9-STAT1 complex
|
0.000607611731082255
|
3
|
77
|
3
|
0.038961038961039
|
1
|
|
GO:BP
|
GO:0045087
|
innate immune response
|
6.61350610150042e-46
|
998
|
126
|
66
|
0.523809523809524
|
0.0661322645290581
|
|
GO:BP
|
GO:0098542
|
defense response to other organism
|
1.35990008321264e-45
|
1218
|
126
|
70
|
0.555555555555556
|
0.0574712643678161
|
|
GO:BP
|
GO:0051707
|
response to other organism
|
7.09190756931387e-44
|
1592
|
126
|
75
|
0.595238095238095
|
0.0471105527638191
|
|
GO:BP
|
GO:0043207
|
response to external biotic stimulus
|
7.09190756931387e-44
|
1594
|
126
|
75
|
0.595238095238095
|
0.0470514429109159
|
|
GO:BP
|
GO:0051607
|
defense response to virus
|
1.41596848889619e-43
|
267
|
143
|
44
|
0.307692307692308
|
0.164794007490637
|
|
GO:BP
|
GO:0140546
|
defense response to symbiont
|
1.41596848889619e-43
|
267
|
143
|
44
|
0.307692307692308
|
0.164794007490637
|
|
GO:BP
|
GO:0009607
|
response to biotic stimulus
|
1.6910333176311e-43
|
1626
|
126
|
75
|
0.595238095238095
|
0.0461254612546125
|
|
GO:BP
|
GO:0009615
|
response to virus
|
7.97093528455231e-42
|
364
|
126
|
45
|
0.357142857142857
|
0.123626373626374
|
|
GO:BP
|
GO:0006955
|
immune response
|
1.89058296515989e-41
|
2311
|
126
|
83
|
0.658730158730159
|
0.0359151882302034
|
|
GO:BP
|
GO:0044419
|
biological process involved in interspecies interaction between organisms
|
4.55133345973674e-40
|
2255
|
126
|
81
|
0.642857142857143
|
0.035920177383592
|
|
GO:BP
|
GO:0006952
|
defense response
|
5.3618746633744e-40
|
1818
|
130
|
76
|
0.584615384615385
|
0.0418041804180418
|
|
GO:BP
|
GO:0060337
|
type I interferon signaling pathway
|
2.69349243769442e-39
|
95
|
117
|
29
|
0.247863247863248
|
0.305263157894737
|
|
GO:BP
|
GO:0071357
|
cellular response to type I interferon
|
3.54565326983405e-39
|
96
|
117
|
29
|
0.247863247863248
|
0.302083333333333
|
|
GO:BP
|
GO:0034340
|
response to type I interferon
|
1.82032307573441e-38
|
101
|
117
|
29
|
0.247863247863248
|
0.287128712871287
|
|
GO:BP
|
GO:0034097
|
response to cytokine
|
4.75883324254128e-37
|
1226
|
85
|
52
|
0.611764705882353
|
0.0424143556280587
|
|
GO:BP
|
GO:0002376
|
immune system process
|
5.305590347357e-32
|
3278
|
126
|
85
|
0.674603174603175
|
0.0259304453935326
|
|
GO:BP
|
GO:0019221
|
cytokine-mediated signaling pathway
|
5.85741883689537e-32
|
810
|
80
|
41
|
0.5125
|
0.0506172839506173
|
|
GO:BP
|
GO:0071345
|
cellular response to cytokine stimulus
|
3.09550120990183e-31
|
1132
|
80
|
45
|
0.5625
|
0.0397526501766784
|
|
GO:BP
|
GO:0034341
|
response to interferon-gamma
|
9.27795325201394e-29
|
196
|
123
|
29
|
0.235772357723577
|
0.147959183673469
|
|
GO:BP
|
GO:0048525
|
negative regulation of viral process
|
4.20882889189609e-28
|
94
|
141
|
24
|
0.170212765957447
|
0.25531914893617
|
|
GO:BP
|
GO:0009605
|
response to external stimulus
|
6.32510879820657e-28
|
2944
|
126
|
77
|
0.611111111111111
|
0.0261548913043478
|
|
GO:BP
|
GO:0045071
|
negative regulation of viral genome replication
|
5.50334954654882e-27
|
54
|
141
|
20
|
0.141843971631206
|
0.37037037037037
|
|
GO:BP
|
GO:0050792
|
regulation of viral process
|
8.41161372508513e-26
|
192
|
141
|
28
|
0.198581560283688
|
0.145833333333333
|
|
GO:BP
|
GO:0002252
|
immune effector process
|
1.03941737639399e-25
|
1309
|
126
|
53
|
0.420634920634921
|
0.040488922841864
|
|
GO:BP
|
GO:0043903
|
regulation of biological process involved in symbiotic interaction
|
3.8737873049691e-25
|
203
|
141
|
28
|
0.198581560283688
|
0.137931034482759
|
|
GO:BP
|
GO:0019079
|
viral genome replication
|
1.95475048975117e-24
|
131
|
141
|
24
|
0.170212765957447
|
0.183206106870229
|
|
GO:BP
|
GO:0045069
|
regulation of viral genome replication
|
2.54140112127626e-24
|
84
|
141
|
21
|
0.148936170212766
|
0.25
|
|
GO:BP
|
GO:0071346
|
cellular response to interferon-gamma
|
5.37886517666107e-23
|
175
|
121
|
24
|
0.198347107438017
|
0.137142857142857
|
|
GO:BP
|
GO:1903900
|
regulation of viral life cycle
|
8.91728361430496e-23
|
153
|
141
|
24
|
0.170212765957447
|
0.156862745098039
|
|
GO:BP
|
GO:0016032
|
viral process
|
2.30992508715859e-22
|
942
|
144
|
46
|
0.319444444444444
|
0.0488322717622081
|
|
GO:BP
|
GO:0044403
|
biological process involved in symbiotic interaction
|
3.27509077469282e-22
|
1001
|
144
|
47
|
0.326388888888889
|
0.046953046953047
|
|
GO:BP
|
GO:0060333
|
interferon-gamma-mediated signaling pathway
|
5.84526715954709e-22
|
88
|
121
|
19
|
0.15702479338843
|
0.215909090909091
|
|
GO:BP
|
GO:0006950
|
response to stress
|
1.62825367021619e-21
|
4170
|
144
|
89
|
0.618055555555556
|
0.0213429256594724
|
|
GO:BP
|
GO:0019058
|
viral life cycle
|
6.96175080212804e-21
|
353
|
141
|
30
|
0.212765957446809
|
0.084985835694051
|
|
GO:BP
|
GO:0071310
|
cellular response to organic substance
|
4.17782362227199e-19
|
2776
|
85
|
51
|
0.6
|
0.018371757925072
|
|
GO:BP
|
GO:0010033
|
response to organic substance
|
1.42443078921624e-18
|
3412
|
80
|
53
|
0.6625
|
0.0155334114888628
|
|
GO:BP
|
GO:0002831
|
regulation of response to biotic stimulus
|
5.12506945127702e-17
|
415
|
126
|
27
|
0.214285714285714
|
0.0650602409638554
|
|
GO:BP
|
GO:0070887
|
cellular response to chemical stimulus
|
1.04212560786315e-16
|
3393
|
99
|
58
|
0.585858585858586
|
0.0170940170940171
|
|
GO:BP
|
GO:0007166
|
cell surface receptor signaling pathway
|
2.47291507274337e-15
|
3210
|
80
|
48
|
0.6
|
0.0149532710280374
|
|
GO:BP
|
GO:0042221
|
response to chemical
|
4.45760210232144e-13
|
4781
|
92
|
60
|
0.652173913043478
|
0.0125496758000418
|
|
GO:BP
|
GO:0035455
|
response to interferon-alpha
|
5.06656957319892e-13
|
23
|
64
|
8
|
0.125
|
0.347826086956522
|
|
GO:BP
|
GO:0031347
|
regulation of defense response
|
3.33990284939831e-12
|
704
|
91
|
24
|
0.263736263736264
|
0.0340909090909091
|
|
GO:BP
|
GO:0045088
|
regulation of innate immune response
|
1.07348614442063e-11
|
309
|
104
|
18
|
0.173076923076923
|
0.058252427184466
|
|
GO:BP
|
GO:0002682
|
regulation of immune system process
|
2.53935125352284e-11
|
1617
|
126
|
40
|
0.317460317460317
|
0.0247371675943105
|
|
GO:BP
|
GO:0035456
|
response to interferon-beta
|
3.76707730303145e-11
|
30
|
133
|
9
|
0.0676691729323308
|
0.3
|
|
GO:BP
|
GO:0002479
|
antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent
|
1.58147427442483e-10
|
75
|
121
|
11
|
0.0909090909090909
|
0.146666666666667
|
|
GO:BP
|
GO:0042590
|
antigen processing and presentation of exogenous peptide antigen via MHC class I
|
3.21967925278366e-10
|
80
|
121
|
11
|
0.0909090909090909
|
0.1375
|
|
GO:BP
|
GO:0032101
|
regulation of response to external stimulus
|
3.36516887986572e-10
|
1051
|
91
|
26
|
0.285714285714286
|
0.0247383444338725
|
|
GO:BP
|
GO:0002474
|
antigen processing and presentation of peptide antigen via MHC class I
|
3.34098436684632e-09
|
99
|
121
|
11
|
0.0909090909090909
|
0.111111111111111
|
|
GO:BP
|
GO:0050776
|
regulation of immune response
|
3.51639631096062e-09
|
996
|
121
|
28
|
0.231404958677686
|
0.0281124497991968
|
|
GO:BP
|
GO:0050896
|
response to stimulus
|
2.03946668175658e-08
|
9407
|
144
|
111
|
0.770833333333333
|
0.0117997236100776
|
|
GO:BP
|
GO:0007165
|
signal transduction
|
2.03946668175658e-08
|
6309
|
91
|
61
|
0.67032967032967
|
0.00966872721508955
|
|
GO:BP
|
GO:0051716
|
cellular response to stimulus
|
2.12928651044164e-08
|
7781
|
92
|
69
|
0.75
|
0.0088677547873024
|
|
GO:BP
|
GO:0023052
|
signaling
|
2.43970140517334e-08
|
6810
|
92
|
64
|
0.695652173913043
|
0.00939794419970631
|
|
GO:BP
|
GO:0048584
|
positive regulation of response to stimulus
|
3.8365268931548e-08
|
2344
|
91
|
35
|
0.384615384615385
|
0.0149317406143345
|
|
GO:BP
|
GO:0002833
|
positive regulation of response to biotic stimulus
|
5.49683202087822e-08
|
250
|
141
|
15
|
0.106382978723404
|
0.06
|
|
GO:BP
|
GO:0080134
|
regulation of response to stress
|
5.61530559100244e-08
|
1443
|
91
|
27
|
0.296703296703297
|
0.0187110187110187
|
|
GO:BP
|
GO:0050688
|
regulation of defense response to virus
|
7.30240208528482e-08
|
71
|
126
|
9
|
0.0714285714285714
|
0.126760563380282
|
|
GO:BP
|
GO:0007154
|
cell communication
|
9.53924872119682e-08
|
6829
|
92
|
63
|
0.684782608695652
|
0.00922536242495241
|
|
GO:BP
|
GO:0002230
|
positive regulation of defense response to virus by host
|
1.16861580070347e-07
|
33
|
126
|
7
|
0.0555555555555556
|
0.212121212121212
|
|
GO:BP
|
GO:0045089
|
positive regulation of innate immune response
|
1.88447772617773e-07
|
216
|
104
|
12
|
0.115384615384615
|
0.0555555555555556
|
|
GO:BP
|
GO:0032479
|
regulation of type I interferon production
|
4.12344683866573e-07
|
130
|
84
|
9
|
0.107142857142857
|
0.0692307692307692
|
|
GO:BP
|
GO:0000209
|
protein polyubiquitination
|
4.27326229819866e-07
|
342
|
140
|
16
|
0.114285714285714
|
0.0467836257309941
|
|
GO:BP
|
GO:0032606
|
type I interferon production
|
4.27326229819866e-07
|
131
|
84
|
9
|
0.107142857142857
|
0.0687022900763359
|
|
GO:BP
|
GO:0050691
|
regulation of defense response to virus by host
|
7.68150528069536e-07
|
43
|
126
|
7
|
0.0555555555555556
|
0.162790697674419
|
|
GO:BP
|
GO:0060700
|
regulation of ribonuclease activity
|
9.08206165432109e-07
|
9
|
67
|
4
|
0.0597014925373134
|
0.444444444444444
|
|
GO:BP
|
GO:0002697
|
regulation of immune effector process
|
9.08206165432109e-07
|
461
|
126
|
17
|
0.134920634920635
|
0.0368763557483731
|
|
GO:BP
|
GO:0002478
|
antigen processing and presentation of exogenous peptide antigen
|
1.00846214528226e-06
|
174
|
121
|
11
|
0.0909090909090909
|
0.0632183908045977
|
|
GO:BP
|
GO:0002684
|
positive regulation of immune system process
|
1.14544239175999e-06
|
1065
|
104
|
23
|
0.221153846153846
|
0.0215962441314554
|
|
GO:BP
|
GO:0019884
|
antigen processing and presentation of exogenous antigen
|
1.55835105738675e-06
|
182
|
121
|
11
|
0.0909090909090909
|
0.0604395604395604
|
|
GO:BP
|
GO:0050778
|
positive regulation of immune response
|
1.98357154476291e-06
|
753
|
104
|
19
|
0.182692307692308
|
0.0252324037184595
|
|
GO:BP
|
GO:0002483
|
antigen processing and presentation of endogenous peptide antigen
|
2.45729711174912e-06
|
19
|
104
|
5
|
0.0480769230769231
|
0.263157894736842
|
|
GO:BP
|
GO:0048002
|
antigen processing and presentation of peptide antigen
|
2.58691028030365e-06
|
192
|
121
|
11
|
0.0909090909090909
|
0.0572916666666667
|
|
GO:BP
|
GO:0032480
|
negative regulation of type I interferon production
|
2.59294769467707e-06
|
46
|
84
|
6
|
0.0714285714285714
|
0.130434782608696
|
|
GO:BP
|
GO:0048583
|
regulation of response to stimulus
|
2.66997361488614e-06
|
4278
|
91
|
45
|
0.494505494505495
|
0.0105189340813464
|
|
GO:BP
|
GO:0001817
|
regulation of cytokine production
|
3.22521936184553e-06
|
806
|
91
|
18
|
0.197802197802198
|
0.0223325062034739
|
|
GO:BP
|
GO:0045824
|
negative regulation of innate immune response
|
3.67819795403632e-06
|
66
|
104
|
7
|
0.0673076923076923
|
0.106060606060606
|
|
GO:BP
|
GO:0001816
|
cytokine production
|
6.25049338414656e-06
|
844
|
91
|
18
|
0.197802197802198
|
0.0213270142180095
|
|
GO:BP
|
GO:0034612
|
response to tumor necrosis factor
|
7.9310902076062e-06
|
331
|
80
|
11
|
0.1375
|
0.0332326283987915
|
|
GO:BP
|
GO:0060760
|
positive regulation of response to cytokine stimulus
|
8.92207230008351e-06
|
57
|
84
|
6
|
0.0714285714285714
|
0.105263157894737
|
|
GO:BP
|
GO:0060759
|
regulation of response to cytokine stimulus
|
1.12758755576927e-05
|
197
|
84
|
9
|
0.107142857142857
|
0.0456852791878173
|
|
GO:BP
|
GO:0006471
|
protein ADP-ribosylation
|
1.12758755576927e-05
|
36
|
141
|
6
|
0.0425531914893617
|
0.166666666666667
|
|
GO:BP
|
GO:0019883
|
antigen processing and presentation of endogenous antigen
|
1.16799734118953e-05
|
26
|
104
|
5
|
0.0480769230769231
|
0.192307692307692
|
|
GO:BP
|
GO:0051336
|
regulation of hydrolase activity
|
1.47082151264892e-05
|
1321
|
91
|
22
|
0.241758241758242
|
0.0166540499621499
|
|
GO:BP
|
GO:0019882
|
antigen processing and presentation
|
1.63235586120204e-05
|
234
|
121
|
11
|
0.0909090909090909
|
0.047008547008547
|
|
GO:BP
|
GO:0046719
|
regulation by virus of viral protein levels in host cell
|
1.70899777352589e-05
|
8
|
39
|
3
|
0.0769230769230769
|
0.375
|
|
GO:BP
|
GO:0032020
|
ISG15-protein conjugation
|
2.03831984046492e-05
|
6
|
58
|
3
|
0.0517241379310345
|
0.5
|
|
GO:BP
|
GO:0048519
|
negative regulation of biological process
|
2.39392996598272e-05
|
5757
|
85
|
49
|
0.576470588235294
|
0.00851137745353483
|
|
GO:BP
|
GO:0061025
|
membrane fusion
|
2.81390916430741e-05
|
179
|
134
|
10
|
0.0746268656716418
|
0.0558659217877095
|
|
GO:BP
|
GO:0071356
|
cellular response to tumor necrosis factor
|
3.01132162822277e-05
|
306
|
80
|
10
|
0.125
|
0.0326797385620915
|
|
GO:BP
|
GO:0031349
|
positive regulation of defense response
|
3.53051550154978e-05
|
362
|
104
|
12
|
0.115384615384615
|
0.0331491712707182
|
|
GO:BP
|
GO:0032069
|
regulation of nuclease activity
|
3.70335958322556e-05
|
22
|
67
|
4
|
0.0597014925373134
|
0.181818181818182
|
|
GO:BP
|
GO:0032103
|
positive regulation of response to external stimulus
|
4.23778549686167e-05
|
513
|
104
|
14
|
0.134615384615385
|
0.0272904483430799
|
|
GO:BP
|
GO:0140289
|
protein mono-ADP-ribosylation
|
4.91506137178473e-05
|
12
|
140
|
4
|
0.0285714285714286
|
0.333333333333333
|
|
GO:BP
|
GO:0002832
|
negative regulation of response to biotic stimulus
|
5.22506824109161e-05
|
100
|
104
|
7
|
0.0673076923076923
|
0.07
|
|
GO:BP
|
GO:0002476
|
antigen processing and presentation of endogenous peptide antigen via MHC class Ib
|
5.44252919924586e-05
|
5
|
104
|
3
|
0.0288461538461538
|
0.6
|
|
GO:BP
|
GO:0035457
|
cellular response to interferon-alpha
|
8.30016378380922e-05
|
13
|
40
|
3
|
0.075
|
0.230769230769231
|
|
GO:BP
|
GO:0050790
|
regulation of catalytic activity
|
9.10017848760616e-05
|
2545
|
86
|
29
|
0.337209302325581
|
0.0113948919449902
|
|
GO:BP
|
GO:0032607
|
interferon-alpha production
|
9.28943660653085e-05
|
30
|
141
|
5
|
0.0354609929078014
|
0.166666666666667
|
|
GO:BP
|
GO:0032647
|
regulation of interferon-alpha production
|
9.28943660653085e-05
|
30
|
141
|
5
|
0.0354609929078014
|
0.166666666666667
|
|
GO:BP
|
GO:1990668
|
vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane
|
9.43212997062539e-05
|
6
|
101
|
3
|
0.0297029702970297
|
0.5
|
|
GO:BP
|
GO:0050777
|
negative regulation of immune response
|
9.52414766605768e-05
|
157
|
104
|
8
|
0.0769230769230769
|
0.0509554140127389
|
|
GO:BP
|
GO:0002428
|
antigen processing and presentation of peptide antigen via MHC class Ib
|
0.000101028910808866
|
6
|
104
|
3
|
0.0288461538461538
|
0.5
|
|
GO:BP
|
GO:0009617
|
response to bacterium
|
0.000112469486224702
|
743
|
91
|
15
|
0.164835164835165
|
0.0201884253028264
|
|
GO:BP
|
GO:0051701
|
biological process involved in interaction with host
|
0.000120961747839942
|
221
|
130
|
10
|
0.0769230769230769
|
0.0452488687782805
|
|
GO:BP
|
GO:0065009
|
regulation of molecular function
|
0.000134294739770239
|
3221
|
86
|
33
|
0.383720930232558
|
0.0102452654455138
|
|
GO:BP
|
GO:0002221
|
pattern recognition receptor signaling pathway
|
0.000173873996513338
|
213
|
141
|
10
|
0.0709219858156028
|
0.0469483568075117
|
|
GO:BP
|
GO:0006954
|
inflammatory response
|
0.000178015876501702
|
794
|
100
|
16
|
0.16
|
0.0201511335012594
|
|
GO:BP
|
GO:0009967
|
positive regulation of signal transduction
|
0.000256888217894991
|
1598
|
91
|
22
|
0.241758241758242
|
0.0137672090112641
|
|
GO:BP
|
GO:0001819
|
positive regulation of cytokine production
|
0.000302912718711211
|
452
|
141
|
14
|
0.099290780141844
|
0.0309734513274336
|
|
GO:BP
|
GO:0052548
|
regulation of endopeptidase activity
|
0.000302912718711211
|
437
|
91
|
11
|
0.120879120879121
|
0.0251716247139588
|
|
GO:BP
|
GO:0033209
|
tumor necrosis factor-mediated signaling pathway
|
0.000330670572862969
|
177
|
80
|
7
|
0.0875
|
0.0395480225988701
|
|
GO:BP
|
GO:0043123
|
positive regulation of I-kappaB kinase/NF-kappaB signaling
|
0.000364232281615712
|
185
|
141
|
9
|
0.0638297872340425
|
0.0486486486486487
|
|
GO:BP
|
GO:0046135
|
pyrimidine nucleoside catabolic process
|
0.000365947519758162
|
22
|
126
|
4
|
0.0317460317460317
|
0.181818181818182
|
|
GO:BP
|
GO:0035458
|
cellular response to interferon-beta
|
0.000369509237445307
|
21
|
133
|
4
|
0.0300751879699248
|
0.19047619047619
|
|
GO:BP
|
GO:0001959
|
regulation of cytokine-mediated signaling pathway
|
0.000378055060587873
|
184
|
79
|
7
|
0.0886075949367089
|
0.0380434782608696
|
|
GO:BP
|
GO:0010647
|
positive regulation of cell communication
|
0.000392364318067873
|
1777
|
91
|
23
|
0.252747252747253
|
0.0129431626336522
|
|
GO:BP
|
GO:0023056
|
positive regulation of signaling
|
0.000407199583537343
|
1782
|
91
|
23
|
0.252747252747253
|
0.0129068462401796
|
|
GO:BP
|
GO:0098586
|
cellular response to virus
|
0.000481930795029178
|
72
|
141
|
6
|
0.0425531914893617
|
0.0833333333333333
|
|
GO:BP
|
GO:0016050
|
vesicle organization
|
0.000481930795029178
|
341
|
101
|
10
|
0.099009900990099
|
0.0293255131964809
|
|
GO:BP
|
GO:1903901
|
negative regulation of viral life cycle
|
0.000481930795029178
|
26
|
115
|
4
|
0.0347826086956522
|
0.153846153846154
|
|
GO:BP
|
GO:0052547
|
regulation of peptidase activity
|
0.000507137333326636
|
467
|
91
|
11
|
0.120879120879121
|
0.0235546038543897
|
|
GO:BP
|
GO:0002718
|
regulation of cytokine production involved in immune response
|
0.000582554530636818
|
85
|
124
|
6
|
0.0483870967741935
|
0.0705882352941176
|
|
GO:BP
|
GO:0043122
|
regulation of I-kappaB kinase/NF-kappaB signaling
|
0.000620117699018922
|
251
|
141
|
10
|
0.0709219858156028
|
0.0398406374501992
|
|
GO:BP
|
GO:0002683
|
negative regulation of immune system process
|
0.000658624973871824
|
420
|
104
|
11
|
0.105769230769231
|
0.0261904761904762
|
|
GO:BP
|
GO:0032727
|
positive regulation of interferon-alpha production
|
0.000735590166149836
|
24
|
141
|
4
|
0.0283687943262411
|
0.166666666666667
|
|
GO:BP
|
GO:0002720
|
positive regulation of cytokine production involved in immune response
|
0.000738689380434938
|
54
|
124
|
5
|
0.0403225806451613
|
0.0925925925925926
|
|
GO:BP
|
GO:0032481
|
positive regulation of type I interferon production
|
0.000760848508813851
|
80
|
84
|
5
|
0.0595238095238095
|
0.0625
|
|
GO:BP
|
GO:0023051
|
regulation of signaling
|
0.000783083757474033
|
3578
|
92
|
35
|
0.380434782608696
|
0.00978200111794299
|
|
GO:BP
|
GO:0052126
|
movement in host environment
|
0.000900701666402316
|
177
|
130
|
8
|
0.0615384615384615
|
0.0451977401129944
|
|
GO:BP
|
GO:0060334
|
regulation of interferon-gamma-mediated signaling pathway
|
0.000923347982380853
|
26
|
48
|
3
|
0.0625
|
0.115384615384615
|
|
GO:BP
|
GO:0060330
|
regulation of response to interferon-gamma
|
0.000923347982380853
|
26
|
48
|
3
|
0.0625
|
0.115384615384615
|
|
GO:BP
|
GO:0019885
|
antigen processing and presentation of endogenous peptide antigen via MHC class I
|
0.000945791852893953
|
13
|
101
|
3
|
0.0297029702970297
|
0.230769230769231
|
|
GO:BP
|
GO:0070555
|
response to interleukin-1
|
0.000945791852893953
|
215
|
80
|
7
|
0.0875
|
0.0325581395348837
|
|
GO:BP
|
GO:0002367
|
cytokine production involved in immune response
|
0.000945791852893953
|
94
|
124
|
6
|
0.0483870967741935
|
0.0638297872340425
|
|
GO:BP
|
GO:0001961
|
positive regulation of cytokine-mediated signaling pathway
|
0.000945791852893953
|
50
|
72
|
4
|
0.0555555555555556
|
0.08
|
|
GO:BP
|
GO:0002475
|
antigen processing and presentation via MHC class Ib
|
0.0010248922848167
|
13
|
104
|
3
|
0.0288461538461538
|
0.230769230769231
|
|
GO:BP
|
GO:0032648
|
regulation of interferon-beta production
|
0.00103988998271197
|
52
|
141
|
5
|
0.0354609929078014
|
0.0961538461538462
|
|
GO:BP
|
GO:0016567
|
protein ubiquitination
|
0.00114956926413069
|
892
|
140
|
19
|
0.135714285714286
|
0.0213004484304933
|
|
GO:BP
|
GO:0032446
|
protein modification by small protein conjugation
|
0.00117533954318843
|
973
|
140
|
20
|
0.142857142857143
|
0.0205549845837616
|
|
GO:BP
|
GO:0032608
|
interferon-beta production
|
0.00122383817909537
|
54
|
141
|
5
|
0.0354609929078014
|
0.0925925925925926
|
|
GO:BP
|
GO:0071222
|
cellular response to lipopolysaccharide
|
0.00130619718166177
|
200
|
91
|
7
|
0.0769230769230769
|
0.035
|
|
GO:BP
|
GO:0001818
|
negative regulation of cytokine production
|
0.00132235530799131
|
381
|
104
|
10
|
0.0961538461538462
|
0.026246719160105
|
|
GO:BP
|
GO:0046718
|
viral entry into host cell
|
0.00137412252638495
|
141
|
130
|
7
|
0.0538461538461538
|
0.049645390070922
|
|
GO:BP
|
GO:0010646
|
regulation of cell communication
|
0.00143446944536411
|
3543
|
92
|
34
|
0.369565217391304
|
0.0095963872424499
|
|
GO:BP
|
GO:0009164
|
nucleoside catabolic process
|
0.00149212320693245
|
33
|
126
|
4
|
0.0317460317460317
|
0.121212121212121
|
|
GO:BP
|
GO:0002730
|
regulation of dendritic cell cytokine production
|
0.0016163399795508
|
13
|
124
|
3
|
0.0241935483870968
|
0.230769230769231
|
|
GO:BP
|
GO:0071360
|
cellular response to exogenous dsRNA
|
0.00166563747506873
|
19
|
84
|
3
|
0.0357142857142857
|
0.157894736842105
|
|
GO:BP
|
GO:0007249
|
I-kappaB kinase/NF-kappaB signaling
|
0.00178038206215671
|
291
|
141
|
10
|
0.0709219858156028
|
0.0343642611683849
|
|
GO:BP
|
GO:0009966
|
regulation of signal transduction
|
0.00181196728954858
|
3165
|
91
|
31
|
0.340659340659341
|
0.00979462875197472
|
|
GO:BP
|
GO:0006213
|
pyrimidine nucleoside metabolic process
|
0.00181196728954858
|
35
|
126
|
4
|
0.0317460317460317
|
0.114285714285714
|
|
GO:BP
|
GO:0071219
|
cellular response to molecule of bacterial origin
|
0.00184424692857634
|
214
|
91
|
7
|
0.0769230769230769
|
0.0327102803738318
|
|
GO:BP
|
GO:0070647
|
protein modification by small protein conjugation or removal
|
0.00194354497659066
|
1184
|
140
|
22
|
0.157142857142857
|
0.0185810810810811
|
|
GO:BP
|
GO:0002371
|
dendritic cell cytokine production
|
0.00194354497659066
|
14
|
124
|
3
|
0.0241935483870968
|
0.214285714285714
|
|
GO:BP
|
GO:0071347
|
cellular response to interleukin-1
|
0.0020676879288766
|
185
|
144
|
8
|
0.0555555555555556
|
0.0432432432432432
|
|
GO:BP
|
GO:0039528
|
cytoplasmic pattern recognition receptor signaling pathway in response to virus
|
0.00208488377106071
|
33
|
141
|
4
|
0.0283687943262411
|
0.121212121212121
|
|
GO:BP
|
GO:0044409
|
entry into host
|
0.00211794030252244
|
154
|
130
|
7
|
0.0538461538461538
|
0.0454545454545455
|
|
GO:BP
|
GO:0072529
|
pyrimidine-containing compound catabolic process
|
0.00233954043845056
|
38
|
126
|
4
|
0.0317460317460317
|
0.105263157894737
|
|
GO:BP
|
GO:0006906
|
vesicle fusion
|
0.00235923902776645
|
108
|
132
|
6
|
0.0454545454545455
|
0.0555555555555556
|
|
GO:BP
|
GO:0009968
|
negative regulation of signal transduction
|
0.00237016830080939
|
1364
|
28
|
9
|
0.321428571428571
|
0.00659824046920821
|
|
GO:BP
|
GO:0071359
|
cellular response to dsRNA
|
0.00265444149122943
|
23
|
84
|
3
|
0.0357142857142857
|
0.130434782608696
|
|
GO:BP
|
GO:0035563
|
positive regulation of chromatin binding
|
0.00266935024842316
|
14
|
142
|
3
|
0.0211267605633803
|
0.214285714285714
|
|
GO:BP
|
GO:0032728
|
positive regulation of interferon-beta production
|
0.00279936680412306
|
36
|
141
|
4
|
0.0283687943262411
|
0.111111111111111
|
|
GO:BP
|
GO:0007005
|
mitochondrion organization
|
0.00279936680412306
|
561
|
39
|
7
|
0.179487179487179
|
0.0124777183600713
|
|
GO:BP
|
GO:0051281
|
positive regulation of release of sequestered calcium ion into cytosol
|
0.00285035841885978
|
42
|
47
|
3
|
0.0638297872340425
|
0.0714285714285714
|
|
GO:BP
|
GO:1901658
|
glycosyl compound catabolic process
|
0.00298100237720974
|
41
|
126
|
4
|
0.0317460317460317
|
0.0975609756097561
|
|
GO:BP
|
GO:0090174
|
organelle membrane fusion
|
0.00299592238824125
|
114
|
132
|
6
|
0.0454545454545455
|
0.0526315789473684
|
|
GO:BP
|
GO:0010522
|
regulation of calcium ion transport into cytosol
|
0.00309832741120827
|
106
|
91
|
5
|
0.0549450549450549
|
0.0471698113207547
|
|
GO:BP
|
GO:0051241
|
negative regulation of multicellular organismal process
|
0.0030985903829046
|
1160
|
91
|
16
|
0.175824175824176
|
0.0137931034482759
|
|
GO:BP
|
GO:0071216
|
cellular response to biotic stimulus
|
0.0030985903829046
|
238
|
91
|
7
|
0.0769230769230769
|
0.0294117647058824
|
|
GO:BP
|
GO:0062207
|
regulation of pattern recognition receptor signaling pathway
|
0.00312284680379508
|
108
|
141
|
6
|
0.0425531914893617
|
0.0555555555555556
|
|
GO:BP
|
GO:0032496
|
response to lipopolysaccharide
|
0.00316970944948925
|
338
|
65
|
7
|
0.107692307692308
|
0.0207100591715976
|
|
GO:BP
|
GO:0044827
|
modulation by host of viral genome replication
|
0.00331738875810053
|
21
|
102
|
3
|
0.0294117647058824
|
0.142857142857143
|
|
GO:BP
|
GO:0038061
|
NIK/NF-kappaB signaling
|
0.00334840715445067
|
194
|
80
|
6
|
0.075
|
0.0309278350515464
|
|
GO:BP
|
GO:0034142
|
toll-like receptor 4 signaling pathway
|
0.00352053133589903
|
44
|
124
|
4
|
0.032258064516129
|
0.0909090909090909
|
|
GO:BP
|
GO:0007259
|
receptor signaling pathway via JAK-STAT
|
0.0036872868475941
|
174
|
91
|
6
|
0.0659340659340659
|
0.0344827586206897
|
|
GO:BP
|
GO:0006508
|
proteolysis
|
0.0036872868475941
|
1836
|
91
|
21
|
0.230769230769231
|
0.011437908496732
|
|
GO:BP
|
GO:0010648
|
negative regulation of cell communication
|
0.00369042272935972
|
1468
|
28
|
9
|
0.321428571428571
|
0.0061307901907357
|
|
GO:BP
|
GO:0002253
|
activation of immune response
|
0.00370668297931851
|
568
|
98
|
11
|
0.112244897959184
|
0.0193661971830986
|
|
GO:BP
|
GO:0023057
|
negative regulation of signaling
|
0.00370893087963302
|
1471
|
28
|
9
|
0.321428571428571
|
0.00611828687967369
|
|
GO:BP
|
GO:0007520
|
myoblast fusion
|
0.00380621708165299
|
42
|
134
|
4
|
0.0298507462686567
|
0.0952380952380952
|
|
GO:BP
|
GO:0070663
|
regulation of leukocyte proliferation
|
0.00394721235762048
|
251
|
120
|
8
|
0.0666666666666667
|
0.0318725099601594
|
|
GO:BP
|
GO:0072527
|
pyrimidine-containing compound metabolic process
|
0.0043737875602442
|
84
|
126
|
5
|
0.0396825396825397
|
0.0595238095238095
|
|
GO:BP
|
GO:0002237
|
response to molecule of bacterial origin
|
0.00441796955953681
|
361
|
65
|
7
|
0.107692307692308
|
0.0193905817174515
|
|
GO:BP
|
GO:0097696
|
receptor signaling pathway via STAT
|
0.004677315022142
|
184
|
91
|
6
|
0.0659340659340659
|
0.0326086956521739
|
|
GO:BP
|
GO:1902036
|
regulation of hematopoietic stem cell differentiation
|
0.00474582724820539
|
75
|
80
|
4
|
0.05
|
0.0533333333333333
|
|
GO:BP
|
GO:0062208
|
positive regulation of pattern recognition receptor signaling pathway
|
0.00477942141477093
|
43
|
141
|
4
|
0.0283687943262411
|
0.0930232558139535
|
|
GO:BP
|
GO:0016553
|
base conversion or substitution editing
|
0.00477942141477093
|
20
|
126
|
3
|
0.0238095238095238
|
0.15
|
|
GO:BP
|
GO:0070098
|
chemokine-mediated signaling pathway
|
0.0049476745048668
|
87
|
70
|
4
|
0.0571428571428571
|
0.0459770114942529
|
|
GO:BP
|
GO:0010950
|
positive regulation of endopeptidase activity
|
0.00495703136033066
|
187
|
91
|
6
|
0.0659340659340659
|
0.0320855614973262
|
|
GO:BP
|
GO:0022603
|
regulation of anatomical structure morphogenesis
|
0.00496855903109591
|
1042
|
18
|
6
|
0.333333333333333
|
0.00575815738963532
|
|
GO:BP
|
GO:0019439
|
aromatic compound catabolic process
|
0.00496855903109591
|
623
|
141
|
14
|
0.099290780141844
|
0.0224719101123595
|
|
GO:BP
|
GO:0034656
|
nucleobase-containing small molecule catabolic process
|
0.00504046733494519
|
49
|
126
|
4
|
0.0317460317460317
|
0.0816326530612245
|
|
GO:BP
|
GO:0000266
|
mitochondrial fission
|
0.00525737156679067
|
41
|
63
|
3
|
0.0476190476190476
|
0.0731707317073171
|
|
GO:BP
|
GO:0008637
|
apoptotic mitochondrial changes
|
0.00536541968516712
|
126
|
21
|
3
|
0.142857142857143
|
0.0238095238095238
|
|
GO:BP
|
GO:0010524
|
positive regulation of calcium ion transport into cytosol
|
0.00551826857686777
|
56
|
47
|
3
|
0.0638297872340425
|
0.0535714285714286
|
|
GO:BP
|
GO:0044766
|
multi-organism transport
|
0.00589082744217206
|
66
|
41
|
3
|
0.0731707317073171
|
0.0454545454545455
|
|
GO:BP
|
GO:1902579
|
multi-organism localization
|
0.00589082744217206
|
66
|
41
|
3
|
0.0731707317073171
|
0.0454545454545455
|
|
GO:BP
|
GO:0034655
|
nucleobase-containing compound catabolic process
|
0.00640426941374343
|
567
|
141
|
13
|
0.0921985815602837
|
0.0229276895943563
|
|
GO:BP
|
GO:0002703
|
regulation of leukocyte mediated immunity
|
0.00675935376917623
|
205
|
124
|
7
|
0.0564516129032258
|
0.0341463414634146
|
|
GO:BP
|
GO:0031348
|
negative regulation of defense response
|
0.00677476080916288
|
246
|
104
|
7
|
0.0673076923076923
|
0.0284552845528455
|
|
GO:BP
|
GO:1990869
|
cellular response to chemokine
|
0.00677476080916288
|
97
|
70
|
4
|
0.0571428571428571
|
0.0412371134020619
|
|
GO:BP
|
GO:1990868
|
response to chemokine
|
0.00677476080916288
|
97
|
70
|
4
|
0.0571428571428571
|
0.0412371134020619
|
|
GO:BP
|
GO:0002520
|
immune system development
|
0.00677476080916288
|
1020
|
91
|
14
|
0.153846153846154
|
0.0137254901960784
|
|
GO:BP
|
GO:0002700
|
regulation of production of molecular mediator of immune response
|
0.00677476080916288
|
148
|
124
|
6
|
0.0483870967741935
|
0.0405405405405405
|
|
GO:BP
|
GO:0019941
|
modification-dependent protein catabolic process
|
0.00677476080916288
|
662
|
29
|
6
|
0.206896551724138
|
0.00906344410876133
|
|
GO:BP
|
GO:1901361
|
organic cyclic compound catabolic process
|
0.00698583690672019
|
651
|
141
|
14
|
0.099290780141844
|
0.021505376344086
|
|
GO:BP
|
GO:0048518
|
positive regulation of biological process
|
0.007129703194716
|
6363
|
106
|
54
|
0.509433962264151
|
0.00848656294200849
|
|
GO:BP
|
GO:0043632
|
modification-dependent macromolecule catabolic process
|
0.0071964284799036
|
673
|
29
|
6
|
0.206896551724138
|
0.00891530460624071
|
|
GO:BP
|
GO:0010952
|
positive regulation of peptidase activity
|
0.0071964284799036
|
205
|
91
|
6
|
0.0659340659340659
|
0.0292682926829268
|
|
GO:BP
|
GO:0048872
|
homeostasis of number of cells
|
0.00720519699234576
|
261
|
12
|
3
|
0.25
|
0.0114942528735632
|
|
GO:BP
|
GO:0002702
|
positive regulation of production of molecular mediator of immune response
|
0.00750918778671368
|
100
|
124
|
5
|
0.0403225806451613
|
0.05
|
|
GO:BP
|
GO:1902533
|
positive regulation of intracellular signal transduction
|
0.00769704061878098
|
1037
|
91
|
14
|
0.153846153846154
|
0.0135004821600771
|
|
GO:BP
|
GO:1901532
|
regulation of hematopoietic progenitor cell differentiation
|
0.00773369780950205
|
89
|
80
|
4
|
0.05
|
0.0449438202247191
|
|
GO:BP
|
GO:0060218
|
hematopoietic stem cell differentiation
|
0.00773369780950205
|
89
|
80
|
4
|
0.05
|
0.0449438202247191
|
|
GO:BP
|
GO:0061844
|
antimicrobial humoral immune response mediated by antimicrobial peptide
|
0.00773486070759078
|
81
|
88
|
4
|
0.0454545454545455
|
0.0493827160493827
|
|
GO:BP
|
GO:0034121
|
regulation of toll-like receptor signaling pathway
|
0.00788786960618308
|
79
|
91
|
4
|
0.043956043956044
|
0.0506329113924051
|
|
GO:BP
|
GO:0030593
|
neutrophil chemotaxis
|
0.00792703595859847
|
103
|
70
|
4
|
0.0571428571428571
|
0.0388349514563107
|
|
GO:BP
|
GO:0002250
|
adaptive immune response
|
0.00810456135297294
|
689
|
121
|
13
|
0.107438016528926
|
0.0188679245283019
|
|
GO:BP
|
GO:0002699
|
positive regulation of immune effector process
|
0.00810456135297294
|
215
|
124
|
7
|
0.0564516129032258
|
0.0325581395348837
|
|
GO:BP
|
GO:0019730
|
antimicrobial humoral response
|
0.00811117891547228
|
145
|
88
|
5
|
0.0568181818181818
|
0.0344827586206897
|
|
GO:BP
|
GO:0030097
|
hemopoiesis
|
0.0082756224134804
|
928
|
91
|
13
|
0.142857142857143
|
0.0140086206896552
|
|
GO:BP
|
GO:0140253
|
cell-cell fusion
|
0.00828996507292287
|
55
|
134
|
4
|
0.0298507462686567
|
0.0727272727272727
|
|
GO:BP
|
GO:0000768
|
syncytium formation by plasma membrane fusion
|
0.00828996507292287
|
55
|
134
|
4
|
0.0298507462686567
|
0.0727272727272727
|
|
GO:BP
|
GO:0010629
|
negative regulation of gene expression
|
0.00849469919458517
|
1460
|
106
|
19
|
0.179245283018868
|
0.013013698630137
|
|
GO:BP
|
GO:0048284
|
organelle fusion
|
0.00849469919458517
|
148
|
132
|
6
|
0.0454545454545455
|
0.0405405405405405
|
|
GO:BP
|
GO:0002218
|
activation of innate immune response
|
0.00890577231645515
|
144
|
91
|
5
|
0.0549450549450549
|
0.0347222222222222
|
|
GO:BP
|
GO:0035561
|
regulation of chromatin binding
|
0.00921877321296962
|
24
|
142
|
3
|
0.0211267605633803
|
0.125
|
|
GO:BP
|
GO:0006949
|
syncytium formation
|
0.00921877321296962
|
57
|
134
|
4
|
0.0298507462686567
|
0.0701754385964912
|
|
GO:BP
|
GO:0044788
|
modulation by host of viral process
|
0.00921877321296962
|
33
|
102
|
3
|
0.0294117647058824
|
0.0909090909090909
|
|
GO:BP
|
GO:0048585
|
negative regulation of response to stimulus
|
0.00921877321296962
|
1733
|
28
|
9
|
0.321428571428571
|
0.0051933064050779
|
|
GO:BP
|
GO:0002224
|
toll-like receptor signaling pathway
|
0.00921877321296962
|
161
|
124
|
6
|
0.0483870967741935
|
0.0372670807453416
|
|
GO:BP
|
GO:0046700
|
heterocycle catabolic process
|
0.00930046199574774
|
602
|
141
|
13
|
0.0921985815602837
|
0.0215946843853821
|
|
GO:BP
|
GO:0002819
|
regulation of adaptive immune response
|
0.00932023493232973
|
167
|
120
|
6
|
0.05
|
0.0359281437125748
|
|
GO:BP
|
GO:0042592
|
homeostatic process
|
0.00938472800021208
|
1981
|
12
|
6
|
0.5
|
0.00302877334679455
|
|
GO:BP
|
GO:0051279
|
regulation of release of sequestered calcium ion into cytosol
|
0.00940195806491978
|
85
|
91
|
4
|
0.043956043956044
|
0.0470588235294118
|
|
GO:BP
|
GO:0035556
|
intracellular signal transduction
|
0.00940195806491978
|
2772
|
91
|
26
|
0.285714285714286
|
0.00937950937950938
|
|
GO:BP
|
GO:2000116
|
regulation of cysteine-type endopeptidase activity
|
0.00974458436501622
|
240
|
85
|
6
|
0.0705882352941176
|
0.025
|
|
GO:BP
|
GO:1904427
|
positive regulation of calcium ion transmembrane transport
|
0.00975492890448954
|
74
|
47
|
3
|
0.0638297872340425
|
0.0405405405405405
|
|
GO:BP
|
GO:0044270
|
cellular nitrogen compound catabolic process
|
0.00977481556511796
|
608
|
141
|
13
|
0.0921985815602837
|
0.0213815789473684
|
|
GO:BP
|
GO:0045595
|
regulation of cell differentiation
|
0.00982681853058317
|
1679
|
6
|
4
|
0.666666666666667
|
0.00238237045860631
|
|
GO:BP
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
0.010098222436342
|
3241
|
117
|
35
|
0.299145299145299
|
0.0107991360691145
|
|
GO:BP
|
GO:0002244
|
hematopoietic progenitor cell differentiation
|
0.010098222436342
|
172
|
80
|
5
|
0.0625
|
0.0290697674418605
|
|
GO:BP
|
GO:0048878
|
chemical homeostasis
|
0.0103215472435808
|
1229
|
47
|
10
|
0.212765957446809
|
0.0081366965012205
|
|
GO:BP
|
GO:0051603
|
proteolysis involved in cellular protein catabolic process
|
0.0104170754822993
|
750
|
29
|
6
|
0.206896551724138
|
0.008
|
|
GO:BP
|
GO:0045596
|
negative regulation of cell differentiation
|
0.0104188477802371
|
697
|
6
|
3
|
0.5
|
0.00430416068866571
|
|
GO:BP
|
GO:0048534
|
hematopoietic or lymphoid organ development
|
0.0104289866910722
|
966
|
91
|
13
|
0.142857142857143
|
0.0134575569358178
|
|
GO:BP
|
GO:0039529
|
RIG-I signaling pathway
|
0.010475204132427
|
26
|
141
|
3
|
0.0212765957446809
|
0.115384615384615
|
|
GO:BP
|
GO:0006900
|
vesicle budding from membrane
|
0.0105595186827448
|
107
|
76
|
4
|
0.0526315789473684
|
0.0373831775700935
|
|
GO:BP
|
GO:0070665
|
positive regulation of leukocyte proliferation
|
0.0108660192355839
|
155
|
91
|
5
|
0.0549450549450549
|
0.032258064516129
|
|
GO:BP
|
GO:0051239
|
regulation of multicellular organismal process
|
0.0108660192355839
|
2861
|
85
|
25
|
0.294117647058824
|
0.00873820342537574
|
|
GO:BP
|
GO:0002429
|
immune response-activating cell surface receptor signaling pathway
|
0.0108884895662746
|
477
|
98
|
9
|
0.0918367346938776
|
0.0188679245283019
|
|
GO:BP
|
GO:0002757
|
immune response-activating signal transduction
|
0.0108884895662746
|
477
|
98
|
9
|
0.0918367346938776
|
0.0188679245283019
|
|
GO:BP
|
GO:0072507
|
divalent inorganic cation homeostasis
|
0.0110478371085169
|
516
|
91
|
9
|
0.0989010989010989
|
0.0174418604651163
|
|
GO:BP
|
GO:0050851
|
antigen receptor-mediated signaling pathway
|
0.0114633098376896
|
323
|
91
|
7
|
0.0769230769230769
|
0.021671826625387
|
|
GO:BP
|
GO:0070661
|
leukocyte proliferation
|
0.0117999952444129
|
325
|
91
|
7
|
0.0769230769230769
|
0.0215384615384615
|
|
GO:BP
|
GO:0006915
|
apoptotic process
|
0.0121648147174474
|
1990
|
95
|
21
|
0.221052631578947
|
0.0105527638190955
|
|
GO:BP
|
GO:0030162
|
regulation of proteolysis
|
0.0124643367367711
|
752
|
91
|
11
|
0.120879120879121
|
0.0146276595744681
|
|
GO:BP
|
GO:1990266
|
neutrophil migration
|
0.0124692956768325
|
124
|
70
|
4
|
0.0571428571428571
|
0.032258064516129
|
|
GO:BP
|
GO:1901224
|
positive regulation of NIK/NF-kappaB signaling
|
0.0129579327380541
|
68
|
59
|
3
|
0.0508474576271186
|
0.0441176470588235
|
|
GO:BP
|
GO:0035567
|
non-canonical Wnt signaling pathway
|
0.013059199223079
|
151
|
27
|
3
|
0.111111111111111
|
0.0198675496688742
|
|
GO:BP
|
GO:0071621
|
granulocyte chemotaxis
|
0.013059199223079
|
126
|
70
|
4
|
0.0571428571428571
|
0.0317460317460317
|
|
GO:BP
|
GO:0060402
|
calcium ion transport into cytosol
|
0.0132431990811777
|
165
|
91
|
5
|
0.0549450549450549
|
0.0303030303030303
|
|
GO:BP
|
GO:0050830
|
defense response to Gram-positive bacterium
|
0.0133649724856615
|
101
|
88
|
4
|
0.0454545454545455
|
0.0396039603960396
|
|
GO:BP
|
GO:0050794
|
regulation of cellular process
|
0.013395583599964
|
11473
|
92
|
72
|
0.782608695652174
|
0.00627560359103983
|
|
GO:BP
|
GO:0002705
|
positive regulation of leukocyte mediated immunity
|
0.0135912556183883
|
122
|
124
|
5
|
0.0403225806451613
|
0.040983606557377
|
|
GO:BP
|
GO:0044257
|
cellular protein catabolic process
|
0.0138856954626843
|
813
|
29
|
6
|
0.206896551724138
|
0.00738007380073801
|
|
GO:BP
|
GO:0002768
|
immune response-regulating cell surface receptor signaling pathway
|
0.0139349465538653
|
512
|
135
|
11
|
0.0814814814814815
|
0.021484375
|
|
GO:BP
|
GO:0043330
|
response to exogenous dsRNA
|
0.0140570060672776
|
50
|
84
|
3
|
0.0357142857142857
|
0.06
|
|
GO:BP
|
GO:0032611
|
interleukin-1 beta production
|
0.0140570060672776
|
107
|
85
|
4
|
0.0470588235294118
|
0.0373831775700935
|
|
GO:BP
|
GO:0002764
|
immune response-regulating signaling pathway
|
0.0142488129982292
|
515
|
135
|
11
|
0.0814814814814815
|
0.0213592233009709
|
|
GO:BP
|
GO:2000736
|
regulation of stem cell differentiation
|
0.0144300573602463
|
115
|
80
|
4
|
0.05
|
0.0347826086956522
|
|
GO:BP
|
GO:0048247
|
lymphocyte chemotaxis
|
0.0158088131148546
|
63
|
70
|
3
|
0.0428571428571429
|
0.0476190476190476
|
|
GO:BP
|
GO:0002440
|
production of molecular mediator of immune response
|
0.0163031447185319
|
189
|
124
|
6
|
0.0483870967741935
|
0.0317460317460317
|
|
GO:BP
|
GO:0042127
|
regulation of cell population proliferation
|
0.016357669110384
|
1774
|
95
|
19
|
0.2
|
0.0107102593010147
|
|
GO:BP
|
GO:0099637
|
neurotransmitter receptor transport
|
0.0166267414128792
|
49
|
92
|
3
|
0.0326086956521739
|
0.0612244897959184
|
|
GO:BP
|
GO:0051246
|
regulation of protein metabolic process
|
0.0166499903138657
|
2704
|
98
|
26
|
0.26530612244898
|
0.00961538461538462
|
|
GO:BP
|
GO:0050852
|
T cell receptor signaling pathway
|
0.0170623127533524
|
203
|
80
|
5
|
0.0625
|
0.0246305418719212
|
|
GO:BP
|
GO:0050789
|
regulation of biological process
|
0.0175841418853843
|
12091
|
93
|
75
|
0.806451612903226
|
0.00620296087999338
|
|
GO:BP
|
GO:0070936
|
protein K48-linked ubiquitination
|
0.0179732249266403
|
61
|
77
|
3
|
0.038961038961039
|
0.0491803278688525
|
|
GO:BP
|
GO:0043331
|
response to dsRNA
|
0.0179804357059627
|
56
|
84
|
3
|
0.0357142857142857
|
0.0535714285714286
|
|
GO:BP
|
GO:0002824
|
positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
|
0.0180893210790963
|
96
|
104
|
4
|
0.0384615384615385
|
0.0416666666666667
|
|
GO:BP
|
GO:0016579
|
protein deubiquitination
|
0.0185714394175951
|
292
|
84
|
6
|
0.0714285714285714
|
0.0205479452054795
|
|
GO:BP
|
GO:0002223
|
stimulatory C-type lectin receptor signaling pathway
|
0.0191186665237093
|
112
|
91
|
4
|
0.043956043956044
|
0.0357142857142857
|
|
GO:BP
|
GO:0060401
|
cytosolic calcium ion transport
|
0.0192004847033772
|
186
|
91
|
5
|
0.0549450549450549
|
0.0268817204301075
|
|
GO:BP
|
GO:0010628
|
positive regulation of gene expression
|
0.0194683847163943
|
1160
|
84
|
13
|
0.154761904761905
|
0.0112068965517241
|
|
GO:BP
|
GO:0002753
|
cytoplasmic pattern recognition receptor signaling pathway
|
0.0194683847163943
|
73
|
141
|
4
|
0.0283687943262411
|
0.0547945205479452
|
|
GO:BP
|
GO:0002886
|
regulation of myeloid leukocyte mediated immunity
|
0.0194683847163943
|
54
|
91
|
3
|
0.032967032967033
|
0.0555555555555556
|
|
GO:BP
|
GO:0033036
|
macromolecule localization
|
0.019577215453504
|
3218
|
102
|
30
|
0.294117647058824
|
0.00932256059664388
|
|
GO:BP
|
GO:0051817
|
modulation of process of other organism involved in symbiotic interaction
|
0.019577215453504
|
102
|
102
|
4
|
0.0392156862745098
|
0.0392156862745098
|
|
GO:BP
|
GO:0055074
|
calcium ion homeostasis
|
0.019577215453504
|
472
|
91
|
8
|
0.0879120879120879
|
0.0169491525423729
|
|
GO:BP
|
GO:0002711
|
positive regulation of T cell mediated immunity
|
0.019577215453504
|
48
|
104
|
3
|
0.0288461538461538
|
0.0625
|
|
GO:BP
|
GO:0002220
|
innate immune response activating cell surface receptor signaling pathway
|
0.0197993037114343
|
115
|
91
|
4
|
0.043956043956044
|
0.0347826086956522
|
|
GO:BP
|
GO:0002821
|
positive regulation of adaptive immune response
|
0.0199773890669071
|
101
|
104
|
4
|
0.0384615384615385
|
0.0396039603960396
|
|
GO:BP
|
GO:0002758
|
innate immune response-activating signal transduction
|
0.0201701195848689
|
116
|
91
|
4
|
0.043956043956044
|
0.0344827586206897
|
|
GO:BP
|
GO:0051093
|
negative regulation of developmental process
|
0.0201701195848689
|
977
|
6
|
3
|
0.5
|
0.00307062436028659
|
|
GO:BP
|
GO:0097530
|
granulocyte migration
|
0.0201701195848689
|
151
|
70
|
4
|
0.0571428571428571
|
0.0264900662251656
|
|
GO:BP
|
GO:0008285
|
negative regulation of cell population proliferation
|
0.0203112871850616
|
790
|
95
|
11
|
0.115789473684211
|
0.0139240506329114
|
|
GO:BP
|
GO:0006521
|
regulation of cellular amino acid metabolic process
|
0.0205009921018207
|
64
|
80
|
3
|
0.0375
|
0.046875
|
|
GO:BP
|
GO:0051702
|
biological process involved in interaction with symbiont
|
0.020617276023941
|
96
|
111
|
4
|
0.036036036036036
|
0.0416666666666667
|
|
GO:BP
|
GO:0015833
|
peptide transport
|
0.0207320836087905
|
1971
|
102
|
21
|
0.205882352941176
|
0.0106544901065449
|
|
GO:BP
|
GO:0032612
|
interleukin-1 production
|
0.0208616171762396
|
126
|
85
|
4
|
0.0470588235294118
|
0.0317460317460317
|
|
GO:BP
|
GO:1902750
|
negative regulation of cell cycle G2/M phase transition
|
0.0209628864320401
|
113
|
95
|
4
|
0.0421052631578947
|
0.0353982300884956
|
|
GO:BP
|
GO:0050727
|
regulation of inflammatory response
|
0.0213989478332364
|
381
|
91
|
7
|
0.0769230769230769
|
0.0183727034120735
|
|
GO:BP
|
GO:0002704
|
negative regulation of leukocyte mediated immunity
|
0.0215943582299139
|
51
|
104
|
3
|
0.0288461538461538
|
0.0588235294117647
|
|
GO:BP
|
GO:0070646
|
protein modification by small protein removal
|
0.0218059747366598
|
310
|
84
|
6
|
0.0714285714285714
|
0.0193548387096774
|
|
GO:BP
|
GO:0140029
|
exocytic process
|
0.0218059747366598
|
83
|
132
|
4
|
0.0303030303030303
|
0.0481927710843374
|
|
GO:BP
|
GO:0012501
|
programmed cell death
|
0.0220055909868952
|
2143
|
95
|
21
|
0.221052631578947
|
0.00979934671021932
|
|
GO:BP
|
GO:0044265
|
cellular macromolecule catabolic process
|
0.0220055909868952
|
1231
|
141
|
19
|
0.134751773049645
|
0.0154346060113729
|
|
GO:BP
|
GO:0016192
|
vesicle-mediated transport
|
0.0221612021545253
|
2180
|
137
|
28
|
0.204379562043796
|
0.0128440366972477
|
|
GO:BP
|
GO:0051651
|
maintenance of location in cell
|
0.0221612021545253
|
223
|
117
|
6
|
0.0512820512820513
|
0.0269058295964126
|
|
GO:BP
|
GO:0008219
|
cell death
|
0.0221612021545253
|
2292
|
95
|
22
|
0.231578947368421
|
0.00959860383944154
|
|
GO:BP
|
GO:0002822
|
regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
|
0.0228216978179719
|
152
|
120
|
5
|
0.0416666666666667
|
0.0328947368421053
|
|
GO:BP
|
GO:0051209
|
release of sequestered calcium ion into cytosol
|
0.0228236225931112
|
123
|
91
|
4
|
0.043956043956044
|
0.032520325203252
|
|
GO:BP
|
GO:0009892
|
negative regulation of metabolic process
|
0.0229421187952566
|
3479
|
117
|
35
|
0.299145299145299
|
0.0100603621730382
|
|
GO:BP
|
GO:0097191
|
extrinsic apoptotic signaling pathway
|
0.0229421187952566
|
223
|
25
|
3
|
0.12
|
0.0134529147982063
|
|
GO:BP
|
GO:0006511
|
ubiquitin-dependent protein catabolic process
|
0.0231196394908779
|
655
|
29
|
5
|
0.172413793103448
|
0.00763358778625954
|
|
GO:BP
|
GO:0043086
|
negative regulation of catalytic activity
|
0.0231196394908779
|
851
|
91
|
11
|
0.120879120879121
|
0.0129259694477086
|
|
GO:BP
|
GO:0072503
|
cellular divalent inorganic cation homeostasis
|
0.0231196394908779
|
497
|
91
|
8
|
0.0879120879120879
|
0.0160965794768612
|
|
GO:BP
|
GO:0051283
|
negative regulation of sequestering of calcium ion
|
0.0231196394908779
|
124
|
91
|
4
|
0.043956043956044
|
0.032258064516129
|
|
GO:BP
|
GO:0042886
|
amide transport
|
0.0233347639798838
|
2007
|
102
|
21
|
0.205882352941176
|
0.0104633781763827
|
|
GO:BP
|
GO:0051282
|
regulation of sequestering of calcium ion
|
0.0239877462772661
|
126
|
91
|
4
|
0.043956043956044
|
0.0317460317460317
|
|
GO:BP
|
GO:0090503
|
RNA phosphodiester bond hydrolysis, exonucleolytic
|
0.0245812295850246
|
43
|
133
|
3
|
0.0225563909774436
|
0.0697674418604651
|
|
GO:BP
|
GO:0030163
|
protein catabolic process
|
0.0247401254270974
|
974
|
29
|
6
|
0.206896551724138
|
0.00616016427104723
|
|
GO:BP
|
GO:0030888
|
regulation of B cell proliferation
|
0.0248396244168403
|
63
|
91
|
3
|
0.032967032967033
|
0.0476190476190476
|
|
GO:BP
|
GO:0030178
|
negative regulation of Wnt signaling pathway
|
0.0255412492506358
|
218
|
27
|
3
|
0.111111111111111
|
0.0137614678899083
|
|
GO:BP
|
GO:0051928
|
positive regulation of calcium ion transport
|
0.0260158836583139
|
125
|
47
|
3
|
0.0638297872340425
|
0.024
|
|
GO:BP
|
GO:0051208
|
sequestering of calcium ion
|
0.0260158836583139
|
130
|
91
|
4
|
0.043956043956044
|
0.0307692307692308
|
|
GO:BP
|
GO:0061024
|
membrane organization
|
0.0262888255159758
|
973
|
143
|
16
|
0.111888111888112
|
0.0164439876670093
|
|
GO:BP
|
GO:0008104
|
protein localization
|
0.0262888255159758
|
2770
|
86
|
23
|
0.267441860465116
|
0.00830324909747292
|
|
GO:BP
|
GO:0050670
|
regulation of lymphocyte proliferation
|
0.0262888255159758
|
230
|
120
|
6
|
0.05
|
0.0260869565217391
|
|
GO:BP
|
GO:0048522
|
positive regulation of cellular process
|
0.0266892030150372
|
5757
|
106
|
47
|
0.443396226415094
|
0.00816397429216606
|
|
GO:BP
|
GO:0050731
|
positive regulation of peptidyl-tyrosine phosphorylation
|
0.0268550975687261
|
198
|
98
|
5
|
0.0510204081632653
|
0.0252525252525253
|
|
GO:BP
|
GO:0032944
|
regulation of mononuclear cell proliferation
|
0.027060762092851
|
232
|
120
|
6
|
0.05
|
0.0258620689655172
|
|
GO:BP
|
GO:0099072
|
regulation of postsynaptic membrane neurotransmitter receptor levels
|
0.02737745551599
|
66
|
92
|
3
|
0.0326086956521739
|
0.0454545454545455
|
|
GO:BP
|
GO:0043462
|
regulation of ATPase activity
|
0.0274238162250587
|
80
|
76
|
3
|
0.0394736842105263
|
0.0375
|
|
GO:BP
|
GO:0002548
|
monocyte chemotaxis
|
0.0275265369626817
|
67
|
91
|
3
|
0.032967032967033
|
0.0447761194029851
|
|
GO:BP
|
GO:0042981
|
regulation of apoptotic process
|
0.0277456284200356
|
1549
|
9
|
4
|
0.444444444444444
|
0.0025823111684958
|
|
GO:BP
|
GO:0071702
|
organic substance transport
|
0.0287255873447741
|
2766
|
102
|
26
|
0.254901960784314
|
0.0093998553868402
|
|
GO:BP
|
GO:0061418
|
regulation of transcription from RNA polymerase II promoter in response to hypoxia
|
0.028914322693781
|
78
|
80
|
3
|
0.0375
|
0.0384615384615385
|
|
GO:BP
|
GO:0050793
|
regulation of developmental process
|
0.0289983857943241
|
2605
|
6
|
4
|
0.666666666666667
|
0.00153550863723608
|
|
GO:BP
|
GO:0045765
|
regulation of angiogenesis
|
0.0289983857943241
|
358
|
18
|
3
|
0.166666666666667
|
0.00837988826815642
|
|
GO:BP
|
GO:0032269
|
negative regulation of cellular protein metabolic process
|
0.0289983857943241
|
1082
|
97
|
13
|
0.134020618556701
|
0.0120147874306839
|
|
GO:BP
|
GO:0015031
|
protein transport
|
0.0290346053697641
|
1929
|
102
|
20
|
0.196078431372549
|
0.0103680663556247
|
|
GO:BP
|
GO:0043067
|
regulation of programmed cell death
|
0.0291533446808788
|
1582
|
9
|
4
|
0.444444444444444
|
0.00252844500632111
|
|
GO:BP
|
GO:0097529
|
myeloid leukocyte migration
|
0.0291533446808788
|
221
|
91
|
5
|
0.0549450549450549
|
0.0226244343891403
|
|
GO:BP
|
GO:0038093
|
Fc receptor signaling pathway
|
0.029498872217318
|
238
|
121
|
6
|
0.0495867768595041
|
0.0252100840336134
|
|
GO:BP
|
GO:1901342
|
regulation of vasculature development
|
0.029498872217318
|
364
|
18
|
3
|
0.166666666666667
|
0.00824175824175824
|
|
GO:BP
|
GO:2001238
|
positive regulation of extrinsic apoptotic signaling pathway
|
0.0298652229623229
|
48
|
135
|
3
|
0.0222222222222222
|
0.0625
|
|
GO:BP
|
GO:0042531
|
positive regulation of tyrosine phosphorylation of STAT protein
|
0.0298652229623229
|
71
|
91
|
3
|
0.032967032967033
|
0.0422535211267606
|
|
GO:BP
|
GO:0065008
|
regulation of biological quality
|
0.0306734134998735
|
4131
|
14
|
8
|
0.571428571428571
|
0.00193657709997579
|
|
GO:BP
|
GO:0033238
|
regulation of cellular amine metabolic process
|
0.0307432994709459
|
82
|
80
|
3
|
0.0375
|
0.0365853658536585
|
|
GO:BP
|
GO:0030595
|
leukocyte chemotaxis
|
0.0307729344150705
|
227
|
91
|
5
|
0.0549450549450549
|
0.0220264317180617
|
|
GO:BP
|
GO:0030099
|
myeloid cell differentiation
|
0.0308528045350106
|
428
|
91
|
7
|
0.0769230769230769
|
0.0163551401869159
|
|
GO:BP
|
GO:0002698
|
negative regulation of immune effector process
|
0.0311225733413905
|
125
|
104
|
4
|
0.0384615384615385
|
0.032
|
|
GO:BP
|
GO:0051851
|
modulation by host of symbiont process
|
0.031149500446077
|
65
|
102
|
3
|
0.0294117647058824
|
0.0461538461538462
|
|
GO:BP
|
GO:0051345
|
positive regulation of hydrolase activity
|
0.031149500446077
|
783
|
91
|
10
|
0.10989010989011
|
0.0127713920817369
|
|
GO:BP
|
GO:0044106
|
cellular amine metabolic process
|
0.031149500446077
|
163
|
80
|
4
|
0.05
|
0.0245398773006135
|
|
GO:BP
|
GO:2001056
|
positive regulation of cysteine-type endopeptidase activity
|
0.0312688191110104
|
154
|
85
|
4
|
0.0470588235294118
|
0.025974025974026
|
|
GO:BP
|
GO:0035821
|
modulation of process of other organism
|
0.0312688191110104
|
128
|
102
|
4
|
0.0392156862745098
|
0.03125
|
|
GO:BP
|
GO:1901222
|
regulation of NIK/NF-kappaB signaling
|
0.0312688191110104
|
113
|
59
|
3
|
0.0508474576271186
|
0.0265486725663717
|
|
GO:BP
|
GO:0007204
|
positive regulation of cytosolic calcium ion concentration
|
0.0312688191110104
|
325
|
91
|
6
|
0.0659340659340659
|
0.0184615384615385
|
|
GO:BP
|
GO:0006401
|
RNA catabolic process
|
0.0313315138824498
|
424
|
141
|
9
|
0.0638297872340425
|
0.0212264150943396
|
|
GO:BP
|
GO:0097553
|
calcium ion transmembrane import into cytosol
|
0.0316757310740357
|
145
|
91
|
4
|
0.043956043956044
|
0.0275862068965517
|
|
GO:BP
|
GO:0031145
|
anaphase-promoting complex-dependent catabolic process
|
0.0316872575923203
|
85
|
80
|
3
|
0.0375
|
0.0352941176470588
|
|
GO:BP
|
GO:0031146
|
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process
|
0.0316872575923203
|
95
|
139
|
4
|
0.0287769784172662
|
0.0421052631578947
|
|
GO:BP
|
GO:0044093
|
positive regulation of molecular function
|
0.0316872575923203
|
1771
|
98
|
18
|
0.183673469387755
|
0.0101637492941841
|
|
GO:BP
|
GO:0055065
|
metal ion homeostasis
|
0.0316872575923203
|
667
|
91
|
9
|
0.0989010989010989
|
0.0134932533733133
|
|
GO:BP
|
GO:0043112
|
receptor metabolic process
|
0.0320117427585339
|
174
|
77
|
4
|
0.051948051948052
|
0.0229885057471264
|
|
GO:BP
|
GO:0061013
|
regulation of mRNA catabolic process
|
0.0321286348228014
|
213
|
141
|
6
|
0.0425531914893617
|
0.028169014084507
|
|
GO:BP
|
GO:0009308
|
amine metabolic process
|
0.0322302804397085
|
168
|
80
|
4
|
0.05
|
0.0238095238095238
|
|
GO:BP
|
GO:0097194
|
execution phase of apoptosis
|
0.0322687197937695
|
96
|
72
|
3
|
0.0416666666666667
|
0.03125
|
|
GO:BP
|
GO:0009116
|
nucleoside metabolic process
|
0.0323462963447996
|
107
|
126
|
4
|
0.0317460317460317
|
0.0373831775700935
|
|
GO:BP
|
GO:0007189
|
adenylate cyclase-activating G protein-coupled receptor signaling pathway
|
0.0324202768428864
|
148
|
47
|
3
|
0.0638297872340425
|
0.0202702702702703
|
|
GO:BP
|
GO:0034504
|
protein localization to nucleus
|
0.032556471628713
|
289
|
138
|
7
|
0.0507246376811594
|
0.0242214532871972
|
|
GO:BP
|
GO:0060341
|
regulation of cellular localization
|
0.0326063636556126
|
851
|
48
|
7
|
0.145833333333333
|
0.0082256169212691
|
|
GO:BP
|
GO:2001236
|
regulation of extrinsic apoptotic signaling pathway
|
0.0327402763852009
|
159
|
135
|
5
|
0.037037037037037
|
0.0314465408805031
|
|
GO:BP
|
GO:0060255
|
regulation of macromolecule metabolic process
|
0.0327708415947404
|
6603
|
86
|
43
|
0.5
|
0.00651219142813872
|
|
GO:BP
|
GO:0010941
|
regulation of cell death
|
0.033085587840949
|
1720
|
9
|
4
|
0.444444444444444
|
0.00232558139534884
|
|
GO:BP
|
GO:1904064
|
positive regulation of cation transmembrane transport
|
0.033085587840949
|
150
|
47
|
3
|
0.0638297872340425
|
0.02
|
|
GO:BP
|
GO:0032268
|
regulation of cellular protein metabolic process
|
0.0333877699363761
|
2541
|
65
|
17
|
0.261538461538462
|
0.00669027941755215
|
|
GO:BP
|
GO:0097190
|
apoptotic signaling pathway
|
0.03365511938105
|
600
|
85
|
8
|
0.0941176470588235
|
0.0133333333333333
|
|
GO:BP
|
GO:0050801
|
ion homeostasis
|
0.0340512181366347
|
808
|
91
|
10
|
0.10989010989011
|
0.0123762376237624
|
|
GO:BP
|
GO:0010951
|
negative regulation of endopeptidase activity
|
0.0340512181366347
|
253
|
55
|
4
|
0.0727272727272727
|
0.0158102766798419
|
|
GO:BP
|
GO:0044248
|
cellular catabolic process
|
0.0342654710789028
|
2347
|
141
|
29
|
0.205673758865248
|
0.0123561994034938
|
|
GO:BP
|
GO:0048523
|
negative regulation of cellular process
|
0.0346455494395674
|
5139
|
85
|
35
|
0.411764705882353
|
0.00681066355322047
|
|
GO:BP
|
GO:0001666
|
response to hypoxia
|
0.035062267473739
|
367
|
85
|
6
|
0.0705882352941176
|
0.0163487738419619
|
|
GO:BP
|
GO:0042267
|
natural killer cell mediated cytotoxicity
|
0.0351825325266614
|
62
|
118
|
3
|
0.0254237288135593
|
0.0483870967741935
|
|
GO:BP
|
GO:0044092
|
negative regulation of molecular function
|
0.0351834233575577
|
1219
|
91
|
13
|
0.142857142857143
|
0.0106644790812141
|
|
GO:BP
|
GO:0002009
|
morphogenesis of an epithelium
|
0.0357275351267642
|
560
|
26
|
4
|
0.153846153846154
|
0.00714285714285714
|
|
GO:BP
|
GO:0001525
|
angiogenesis
|
0.0359570848178843
|
617
|
37
|
5
|
0.135135135135135
|
0.00810372771474878
|
|
GO:BP
|
GO:2000779
|
regulation of double-strand break repair
|
0.0359570848178843
|
87
|
85
|
3
|
0.0352941176470588
|
0.0344827586206897
|
|
GO:BP
|
GO:1903169
|
regulation of calcium ion transmembrane transport
|
0.0359570848178843
|
158
|
47
|
3
|
0.0638297872340425
|
0.0189873417721519
|
|
GO:BP
|
GO:0032760
|
positive regulation of tumor necrosis factor production
|
0.0359570848178843
|
88
|
84
|
3
|
0.0357142857142857
|
0.0340909090909091
|
|
GO:BP
|
GO:0032102
|
negative regulation of response to external stimulus
|
0.0361386964350436
|
406
|
78
|
6
|
0.0769230769230769
|
0.0147783251231527
|
|
GO:BP
|
GO:0051248
|
negative regulation of protein metabolic process
|
0.0368177047909823
|
1151
|
97
|
13
|
0.134020618556701
|
0.0112945264986968
|
|
GO:BP
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
0.0368177047909823
|
3483
|
106
|
31
|
0.292452830188679
|
0.00890037324145851
|
|
GO:BP
|
GO:0006874
|
cellular calcium ion homeostasis
|
0.0370442170246945
|
460
|
91
|
7
|
0.0769230769230769
|
0.0152173913043478
|
|
GO:BP
|
GO:2000026
|
regulation of multicellular organismal development
|
0.0370442170246945
|
1447
|
6
|
3
|
0.5
|
0.00207325501036628
|
|
GO:BP
|
GO:0009888
|
tissue development
|
0.0372746433475759
|
2107
|
8
|
4
|
0.5
|
0.0018984337921215
|
|
GO:BP
|
GO:0006820
|
anion transport
|
0.0372951283031747
|
2889
|
102
|
26
|
0.254901960784314
|
0.00899965385946694
|
|
GO:BP
|
GO:1903557
|
positive regulation of tumor necrosis factor superfamily cytokine production
|
0.0373389429847414
|
91
|
84
|
3
|
0.0357142857142857
|
0.032967032967033
|
|
GO:BP
|
GO:0010466
|
negative regulation of peptidase activity
|
0.0373648909317498
|
267
|
55
|
4
|
0.0727272727272727
|
0.0149812734082397
|
|
GO:BP
|
GO:0002228
|
natural killer cell mediated immunity
|
0.0374348409544926
|
65
|
118
|
3
|
0.0254237288135593
|
0.0461538461538462
|
|
GO:BP
|
GO:0002709
|
regulation of T cell mediated immunity
|
0.0377287523769019
|
74
|
104
|
3
|
0.0288461538461538
|
0.0405405405405405
|
|
GO:BP
|
GO:0043085
|
positive regulation of catalytic activity
|
0.0380558680699972
|
1418
|
98
|
15
|
0.153061224489796
|
0.0105782792665726
|
|
GO:BP
|
GO:0045184
|
establishment of protein localization
|
0.0380558680699972
|
2035
|
102
|
20
|
0.196078431372549
|
0.00982800982800983
|
|
GO:BP
|
GO:0036293
|
response to decreased oxygen levels
|
0.0382947095681582
|
382
|
85
|
6
|
0.0705882352941176
|
0.0157068062827225
|
|
GO:BP
|
GO:0071705
|
nitrogen compound transport
|
0.0385106988327389
|
2322
|
102
|
22
|
0.215686274509804
|
0.00947459086993971
|
|
GO:BP
|
GO:0006919
|
activation of cysteine-type endopeptidase activity involved in apoptotic process
|
0.0385439812686909
|
92
|
85
|
3
|
0.0352941176470588
|
0.0326086956521739
|
|
GO:BP
|
GO:0008283
|
cell population proliferation
|
0.0394538176871928
|
2054
|
95
|
19
|
0.2
|
0.00925024342745862
|
|
GO:BP
|
GO:0051480
|
regulation of cytosolic calcium ion concentration
|
0.0394538176871928
|
360
|
91
|
6
|
0.0659340659340659
|
0.0166666666666667
|
|
GO:BP
|
GO:0010468
|
regulation of gene expression
|
0.0394538176871928
|
5196
|
97
|
39
|
0.402061855670103
|
0.00750577367205543
|
|
GO:BP
|
GO:0042509
|
regulation of tyrosine phosphorylation of STAT protein
|
0.0394975737545814
|
87
|
91
|
3
|
0.032967032967033
|
0.0344827586206897
|
|
GO:BP
|
GO:1902531
|
regulation of intracellular signal transduction
|
0.0395903589501495
|
1842
|
91
|
17
|
0.186813186813187
|
0.00922909880564604
|
|
GO:BP
|
GO:0042157
|
lipoprotein metabolic process
|
0.0399030761725486
|
136
|
111
|
4
|
0.036036036036036
|
0.0294117647058824
|
|
GO:BP
|
GO:0006875
|
cellular metal ion homeostasis
|
0.0399030761725486
|
593
|
91
|
8
|
0.0879120879120879
|
0.0134907251264755
|
|
GO:BP
|
GO:0070498
|
interleukin-1-mediated signaling pathway
|
0.0399030761725486
|
105
|
144
|
4
|
0.0277777777777778
|
0.0380952380952381
|
|
GO:BP
|
GO:0048646
|
anatomical structure formation involved in morphogenesis
|
0.0399030761725486
|
1202
|
37
|
7
|
0.189189189189189
|
0.00582362728785358
|
|
GO:BP
|
GO:0051924
|
regulation of calcium ion transport
|
0.0399030761725486
|
259
|
91
|
5
|
0.0549450549450549
|
0.0193050193050193
|
|
GO:BP
|
GO:0038095
|
Fc-epsilon receptor signaling pathway
|
0.0402196760009331
|
167
|
91
|
4
|
0.043956043956044
|
0.0239520958083832
|
|
GO:BP
|
GO:0070304
|
positive regulation of stress-activated protein kinase signaling cascade
|
0.0405812182854595
|
123
|
124
|
4
|
0.032258064516129
|
0.032520325203252
|
|
GO:BP
|
GO:0007260
|
tyrosine phosphorylation of STAT protein
|
0.0406151718894078
|
89
|
91
|
3
|
0.032967032967033
|
0.0337078651685393
|
|
GO:BP
|
GO:0072676
|
lymphocyte migration
|
0.0413394578927084
|
117
|
70
|
3
|
0.0428571428571429
|
0.0256410256410256
|
|
GO:BP
|
GO:0065007
|
biological regulation
|
0.0413826614271488
|
12809
|
93
|
76
|
0.817204301075269
|
0.00593332812865954
|
|
GO:BP
|
GO:0006959
|
humoral immune response
|
0.0426954661833603
|
384
|
88
|
6
|
0.0681818181818182
|
0.015625
|
|
GO:BP
|
GO:0010959
|
regulation of metal ion transport
|
0.0434592394756691
|
268
|
91
|
5
|
0.0549450549450549
|
0.0186567164179104
|
|
GO:BP
|
GO:0051100
|
negative regulation of binding
|
0.0438800161874172
|
163
|
52
|
3
|
0.0576923076923077
|
0.0184049079754601
|
|
GO:BP
|
GO:0010803
|
regulation of tumor necrosis factor-mediated signaling pathway
|
0.0441819917996985
|
63
|
135
|
3
|
0.0222222222222222
|
0.0476190476190476
|
|
GO:BP
|
GO:0014902
|
myotube differentiation
|
0.044791957407179
|
119
|
134
|
4
|
0.0298507462686567
|
0.0336134453781513
|
|
GO:BP
|
GO:0045862
|
positive regulation of proteolysis
|
0.0452970276995125
|
378
|
91
|
6
|
0.0659340659340659
|
0.0158730158730159
|
|
GO:BP
|
GO:0032651
|
regulation of interleukin-1 beta production
|
0.0460684077881814
|
102
|
85
|
3
|
0.0352941176470588
|
0.0294117647058824
|
|
GO:BP
|
GO:0070482
|
response to oxygen levels
|
0.0461963086969165
|
408
|
85
|
6
|
0.0705882352941176
|
0.0147058823529412
|
|
GO:BP
|
GO:0006520
|
cellular amino acid metabolic process
|
0.04642330414943
|
341
|
26
|
3
|
0.115384615384615
|
0.00879765395894428
|
|
GO:BP
|
GO:0009057
|
macromolecule catabolic process
|
0.0465479134443224
|
1468
|
32
|
7
|
0.21875
|
0.00476839237057221
|
|
GO:BP
|
GO:1901564
|
organonitrogen compound metabolic process
|
0.0472298653367356
|
6792
|
112
|
54
|
0.482142857142857
|
0.00795053003533569
|
|
GO:BP
|
GO:1901657
|
glycosyl compound metabolic process
|
0.0472301407583929
|
130
|
126
|
4
|
0.0317460317460317
|
0.0307692307692308
|
|
GO:BP
|
GO:0055080
|
cation homeostasis
|
0.0472504104558002
|
750
|
91
|
9
|
0.0989010989010989
|
0.012
|
|
GO:BP
|
GO:0019222
|
regulation of metabolic process
|
0.0475092008361065
|
7148
|
98
|
50
|
0.510204081632653
|
0.00699496362618914
|
|
GO:BP
|
GO:0042100
|
B cell proliferation
|
0.0478824524424826
|
98
|
91
|
3
|
0.032967032967033
|
0.0306122448979592
|
|
GO:BP
|
GO:0048729
|
tissue morphogenesis
|
0.0481378100950546
|
667
|
14
|
3
|
0.214285714285714
|
0.00449775112443778
|
|
GO:BP
|
GO:0051240
|
positive regulation of multicellular organismal process
|
0.0484552005135169
|
1455
|
91
|
14
|
0.153846153846154
|
0.00962199312714777
|
|
GO:BP
|
GO:0046651
|
lymphocyte proliferation
|
0.0487113908520208
|
294
|
120
|
6
|
0.05
|
0.0204081632653061
|
|
GO:BP
|
GO:1901701
|
cellular response to oxygen-containing compound
|
0.0488797046592785
|
1227
|
97
|
13
|
0.134020618556701
|
0.0105949470252649
|
|
GO:BP
|
GO:0051346
|
negative regulation of hydrolase activity
|
0.0493579887017317
|
470
|
55
|
5
|
0.0909090909090909
|
0.0106382978723404
|
|
GO:CC
|
GO:0005829
|
cytosol
|
7.66315904181968e-08
|
5303
|
97
|
57
|
0.587628865979381
|
0.0107486328493306
|
|
GO:CC
|
GO:0005737
|
cytoplasm
|
2.89034781025271e-07
|
11911
|
144
|
123
|
0.854166666666667
|
0.0103265888674335
|
|
GO:CC
|
GO:0030670
|
phagocytic vesicle membrane
|
0.000203175585383269
|
76
|
104
|
6
|
0.0576923076923077
|
0.0789473684210526
|
|
GO:CC
|
GO:0097708
|
intracellular vesicle
|
0.000203175585383269
|
2428
|
137
|
38
|
0.277372262773723
|
0.0156507413509061
|
|
GO:CC
|
GO:0031410
|
cytoplasmic vesicle
|
0.000203175585383269
|
2424
|
137
|
38
|
0.277372262773723
|
0.0156765676567657
|
|
GO:CC
|
GO:0012505
|
endomembrane system
|
0.000203175585383269
|
4640
|
140
|
59
|
0.421428571428571
|
0.0127155172413793
|
|
GO:CC
|
GO:0045335
|
phagocytic vesicle
|
0.000203175585383269
|
137
|
124
|
8
|
0.0645161290322581
|
0.0583941605839416
|
|
GO:CC
|
GO:0030666
|
endocytic vesicle membrane
|
0.000389837261156929
|
159
|
121
|
8
|
0.0661157024793388
|
0.050314465408805
|
|
GO:CC
|
GO:0042824
|
MHC class I peptide loading complex
|
0.000453955979869482
|
9
|
101
|
3
|
0.0297029702970297
|
0.333333333333333
|
|
GO:CC
|
GO:0048471
|
perinuclear region of cytoplasm
|
0.000453955979869482
|
735
|
58
|
11
|
0.189655172413793
|
0.0149659863945578
|
|
GO:CC
|
GO:0030139
|
endocytic vesicle
|
0.00104806784075597
|
306
|
124
|
10
|
0.0806451612903226
|
0.0326797385620915
|
|
GO:CC
|
GO:0031982
|
vesicle
|
0.00116731932753578
|
4055
|
138
|
50
|
0.36231884057971
|
0.0123304562268804
|
|
GO:CC
|
GO:0005794
|
Golgi apparatus
|
0.00168178904209921
|
1623
|
105
|
22
|
0.20952380952381
|
0.0135551447935921
|
|
GO:CC
|
GO:0005622
|
intracellular anatomical structure
|
0.00191778851520003
|
14790
|
144
|
130
|
0.902777777777778
|
0.00878972278566599
|
|
GO:CC
|
GO:0016605
|
PML body
|
0.00195620616662585
|
102
|
136
|
6
|
0.0441176470588235
|
0.0588235294117647
|
|
GO:CC
|
GO:0043231
|
intracellular membrane-bounded organelle
|
0.00379996180158052
|
11296
|
142
|
105
|
0.73943661971831
|
0.00929532577903683
|
|
GO:CC
|
GO:0071556
|
integral component of lumenal side of endoplasmic reticulum membrane
|
0.00489729964615974
|
25
|
104
|
3
|
0.0288461538461538
|
0.12
|
|
GO:CC
|
GO:0098553
|
lumenal side of endoplasmic reticulum membrane
|
0.00489729964615974
|
25
|
104
|
3
|
0.0288461538461538
|
0.12
|
|
GO:CC
|
GO:0012506
|
vesicle membrane
|
0.00714934564531732
|
800
|
134
|
15
|
0.111940298507463
|
0.01875
|
|
GO:CC
|
GO:0031901
|
early endosome membrane
|
0.00714934564531732
|
159
|
121
|
6
|
0.0495867768595041
|
0.0377358490566038
|
|
GO:CC
|
GO:0098576
|
lumenal side of membrane
|
0.00943811539232163
|
33
|
104
|
3
|
0.0288461538461538
|
0.0909090909090909
|
|
GO:CC
|
GO:0005768
|
endosome
|
0.00943811539232163
|
987
|
137
|
17
|
0.124087591240876
|
0.0172239108409321
|
|
GO:CC
|
GO:0005769
|
early endosome
|
0.0130544664042596
|
378
|
130
|
9
|
0.0692307692307692
|
0.0238095238095238
|
|
GO:CC
|
GO:0030659
|
cytoplasmic vesicle membrane
|
0.0130544664042596
|
779
|
134
|
14
|
0.104477611940299
|
0.0179717586649551
|
|
GO:CC
|
GO:0043227
|
membrane-bounded organelle
|
0.0130544664042596
|
12858
|
142
|
113
|
0.795774647887324
|
0.00878830300202209
|
|
GO:CC
|
GO:0010008
|
endosome membrane
|
0.0151445557325766
|
505
|
121
|
10
|
0.0826446280991736
|
0.0198019801980198
|
|
GO:CC
|
GO:0043229
|
intracellular organelle
|
0.0168477525823618
|
12491
|
142
|
110
|
0.774647887323944
|
0.00880634056520695
|
|
GO:CC
|
GO:0030176
|
integral component of endoplasmic reticulum membrane
|
0.0168477525823618
|
158
|
104
|
5
|
0.0480769230769231
|
0.0316455696202532
|
|
GO:CC
|
GO:0005576
|
extracellular region
|
0.0179304386567665
|
4567
|
92
|
35
|
0.380434782608696
|
0.0076636741843661
|
|
GO:CC
|
GO:0098588
|
bounding membrane of organelle
|
0.0185706079621181
|
2158
|
127
|
26
|
0.204724409448819
|
0.0120481927710843
|
|
GO:CC
|
GO:0005634
|
nucleus
|
0.0190026782800649
|
7592
|
144
|
75
|
0.520833333333333
|
0.00987881981032666
|
|
GO:CC
|
GO:0031227
|
intrinsic component of endoplasmic reticulum membrane
|
0.0190026782800649
|
166
|
104
|
5
|
0.0480769230769231
|
0.0301204819277108
|
|
GO:CC
|
GO:0000502
|
proteasome complex
|
0.0224070923441574
|
66
|
80
|
3
|
0.0375
|
0.0454545454545455
|
|
GO:CC
|
GO:1905369
|
endopeptidase complex
|
0.0280694638545862
|
72
|
80
|
3
|
0.0375
|
0.0416666666666667
|
|
GO:CC
|
GO:0031981
|
nuclear lumen
|
0.0386787741753497
|
4444
|
69
|
26
|
0.376811594202899
|
0.00585058505850585
|
|
GO:CC
|
GO:0031090
|
organelle membrane
|
0.0386787741753497
|
3612
|
104
|
31
|
0.298076923076923
|
0.00858250276854928
|
|
GO:CC
|
GO:0043226
|
organelle
|
0.0411777378302528
|
14008
|
142
|
118
|
0.830985915492958
|
0.00842375785265563
|
|
GO:CC
|
GO:0031965
|
nuclear membrane
|
0.0411777378302528
|
298
|
138
|
7
|
0.0507246376811594
|
0.023489932885906
|
|
GO:CC
|
GO:0005739
|
mitochondrion
|
0.0434696811946228
|
1673
|
40
|
9
|
0.225
|
0.00537955768081291
|
|
GO:CC
|
GO:0005783
|
endoplasmic reticulum
|
0.0435629911352729
|
1982
|
144
|
25
|
0.173611111111111
|
0.0126135216952573
|
|
GO:CC
|
GO:0031984
|
organelle subcompartment
|
0.0435629911352729
|
1773
|
104
|
18
|
0.173076923076923
|
0.0101522842639594
|
|
GO:CC
|
GO:0033116
|
endoplasmic reticulum-Golgi intermediate compartment membrane
|
0.0435629911352729
|
74
|
101
|
3
|
0.0297029702970297
|
0.0405405405405405
|
|
GO:CC
|
GO:0005654
|
nucleoplasm
|
0.0462719518782395
|
4102
|
69
|
24
|
0.347826086956522
|
0.00585080448561677
|
|
GO:MF
|
GO:0003725
|
double-stranded RNA binding
|
3.30497046500695e-08
|
77
|
90
|
9
|
0.1
|
0.116883116883117
|
|
GO:MF
|
GO:0001730
|
2'-5'-oligoadenylate synthetase activity
|
3.30497046500695e-08
|
4
|
67
|
4
|
0.0597014925373134
|
1
|
|
GO:MF
|
GO:0042802
|
identical protein binding
|
9.26264558001886e-07
|
2067
|
138
|
40
|
0.289855072463768
|
0.019351717464925
|
|
GO:MF
|
GO:0046978
|
TAP1 binding
|
1.47427901080936e-06
|
6
|
104
|
4
|
0.0384615384615385
|
0.666666666666667
|
|
GO:MF
|
GO:0046977
|
TAP binding
|
9.7804095481826e-06
|
9
|
104
|
4
|
0.0384615384615385
|
0.444444444444444
|
|
GO:MF
|
GO:0048248
|
CXCR3 chemokine receptor binding
|
1.04730289131977e-05
|
5
|
47
|
3
|
0.0638297872340425
|
0.6
|
|
GO:MF
|
GO:0046979
|
TAP2 binding
|
1.04730289131977e-05
|
3
|
104
|
3
|
0.0288461538461538
|
1
|
|
GO:MF
|
GO:0042605
|
peptide antigen binding
|
3.38685445756901e-05
|
30
|
104
|
5
|
0.0480769230769231
|
0.166666666666667
|
|
GO:MF
|
GO:0042379
|
chemokine receptor binding
|
5.444488654301e-05
|
72
|
88
|
6
|
0.0681818181818182
|
0.0833333333333333
|
|
GO:MF
|
GO:0005525
|
GTP binding
|
5.51670015998935e-05
|
381
|
56
|
9
|
0.160714285714286
|
0.0236220472440945
|
|
GO:MF
|
GO:0097367
|
carbohydrate derivative binding
|
5.51670015998935e-05
|
2285
|
57
|
21
|
0.368421052631579
|
0.00919037199124726
|
|
GO:MF
|
GO:0015440
|
ABC-type peptide transporter activity
|
5.51670015998935e-05
|
5
|
101
|
3
|
0.0297029702970297
|
0.6
|
|
GO:MF
|
GO:0032549
|
ribonucleoside binding
|
5.51670015998935e-05
|
389
|
56
|
9
|
0.160714285714286
|
0.0231362467866324
|
|
GO:MF
|
GO:0032550
|
purine ribonucleoside binding
|
5.51670015998935e-05
|
386
|
56
|
9
|
0.160714285714286
|
0.0233160621761658
|
|
GO:MF
|
GO:0003950
|
NAD+ ADP-ribosyltransferase activity
|
5.51670015998935e-05
|
27
|
141
|
5
|
0.0354609929078014
|
0.185185185185185
|
|
GO:MF
|
GO:0001883
|
purine nucleoside binding
|
5.51670015998935e-05
|
389
|
56
|
9
|
0.160714285714286
|
0.0231362467866324
|
|
GO:MF
|
GO:0015433
|
ABC-type peptide antigen transporter activity
|
5.51670015998935e-05
|
5
|
101
|
3
|
0.0297029702970297
|
0.6
|
|
GO:MF
|
GO:0001882
|
nucleoside binding
|
5.83211797686557e-05
|
395
|
56
|
9
|
0.160714285714286
|
0.0227848101265823
|
|
GO:MF
|
GO:0017111
|
nucleoside-triphosphatase activity
|
5.83211797686557e-05
|
872
|
119
|
19
|
0.159663865546218
|
0.0217889908256881
|
|
GO:MF
|
GO:0019001
|
guanyl nucleotide binding
|
5.96491416105801e-05
|
403
|
56
|
9
|
0.160714285714286
|
0.0223325062034739
|
|
GO:MF
|
GO:0032561
|
guanyl ribonucleotide binding
|
5.96491416105801e-05
|
403
|
56
|
9
|
0.160714285714286
|
0.0223325062034739
|
|
GO:MF
|
GO:0005164
|
tumor necrosis factor receptor binding
|
6.07203445258867e-05
|
32
|
17
|
3
|
0.176470588235294
|
0.09375
|
|
GO:MF
|
GO:0042288
|
MHC class I protein binding
|
6.93942847188183e-05
|
20
|
101
|
4
|
0.0396039603960396
|
0.2
|
|
GO:MF
|
GO:0016763
|
transferase activity, transferring pentosyl groups
|
7.21371815086572e-05
|
56
|
141
|
6
|
0.0425531914893617
|
0.107142857142857
|
|
GO:MF
|
GO:0042803
|
protein homodimerization activity
|
7.59494880701919e-05
|
676
|
56
|
11
|
0.196428571428571
|
0.0162721893491124
|
|
GO:MF
|
GO:1990404
|
protein ADP-ribosylase activity
|
8.62787492579643e-05
|
16
|
140
|
4
|
0.0285714285714286
|
0.25
|
|
GO:MF
|
GO:0035639
|
purine ribonucleoside triphosphate binding
|
8.70492803676134e-05
|
1847
|
57
|
18
|
0.315789473684211
|
0.00974553329723877
|
|
GO:MF
|
GO:0070566
|
adenylyltransferase activity
|
8.93236727716187e-05
|
33
|
67
|
4
|
0.0597014925373134
|
0.121212121212121
|
|
GO:MF
|
GO:0016818
|
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
|
9.06510974633053e-05
|
930
|
119
|
19
|
0.159663865546218
|
0.0204301075268817
|
|
GO:MF
|
GO:0016462
|
pyrophosphatase activity
|
9.06510974633053e-05
|
928
|
119
|
19
|
0.159663865546218
|
0.0204741379310345
|
|
GO:MF
|
GO:0016817
|
hydrolase activity, acting on acid anhydrides
|
9.06510974633053e-05
|
930
|
119
|
19
|
0.159663865546218
|
0.0204301075268817
|
|
GO:MF
|
GO:0032555
|
purine ribonucleotide binding
|
0.000124641940745394
|
1918
|
57
|
18
|
0.315789473684211
|
0.00938477580813347
|
|
GO:MF
|
GO:0017076
|
purine nucleotide binding
|
0.000132636802197205
|
1933
|
57
|
18
|
0.315789473684211
|
0.00931195033626487
|
|
GO:MF
|
GO:0032553
|
ribonucleotide binding
|
0.000132636802197205
|
1935
|
57
|
18
|
0.315789473684211
|
0.00930232558139535
|
|
GO:MF
|
GO:0005515
|
protein binding
|
0.000136522163403669
|
14767
|
106
|
100
|
0.943396226415094
|
0.00677185616577504
|
|
GO:MF
|
GO:0032813
|
tumor necrosis factor receptor superfamily binding
|
0.000136522163403669
|
49
|
17
|
3
|
0.176470588235294
|
0.0612244897959184
|
|
GO:MF
|
GO:0045236
|
CXCR chemokine receptor binding
|
0.000140197252352119
|
18
|
47
|
3
|
0.0638297872340425
|
0.166666666666667
|
|
GO:MF
|
GO:0044389
|
ubiquitin-like protein ligase binding
|
0.000257715819720571
|
314
|
91
|
9
|
0.0989010989010989
|
0.0286624203821656
|
|
GO:MF
|
GO:0000166
|
nucleotide binding
|
0.000324049193231373
|
2171
|
60
|
19
|
0.316666666666667
|
0.00875172731460157
|
|
GO:MF
|
GO:1901265
|
nucleoside phosphate binding
|
0.000324049193231373
|
2172
|
60
|
19
|
0.316666666666667
|
0.00874769797421731
|
|
GO:MF
|
GO:0008009
|
chemokine activity
|
0.000359802053798525
|
49
|
70
|
4
|
0.0571428571428571
|
0.0816326530612245
|
|
GO:MF
|
GO:0005126
|
cytokine receptor binding
|
0.000482318026467555
|
275
|
88
|
8
|
0.0909090909090909
|
0.0290909090909091
|
|
GO:MF
|
GO:0003924
|
GTPase activity
|
0.000544483859880313
|
328
|
56
|
7
|
0.125
|
0.0213414634146341
|
|
GO:MF
|
GO:0042287
|
MHC protein binding
|
0.00055646149048061
|
39
|
101
|
4
|
0.0396039603960396
|
0.102564102564103
|
|
GO:MF
|
GO:0043168
|
anion binding
|
0.000564100184129994
|
2418
|
57
|
19
|
0.333333333333333
|
0.00785773366418528
|
|
GO:MF
|
GO:0005125
|
cytokine activity
|
0.000669613867228356
|
235
|
81
|
7
|
0.0864197530864197
|
0.0297872340425532
|
|
GO:MF
|
GO:0016787
|
hydrolase activity
|
0.000795573456344697
|
2615
|
101
|
29
|
0.287128712871287
|
0.0110898661567878
|
|
GO:MF
|
GO:0016814
|
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines
|
0.000795573456344697
|
35
|
126
|
4
|
0.0317460317460317
|
0.114285714285714
|
|
GO:MF
|
GO:0098772
|
molecular function regulator
|
0.00103308277301778
|
2363
|
86
|
24
|
0.27906976744186
|
0.0101565806178587
|
|
GO:MF
|
GO:0005488
|
binding
|
0.00167116303050046
|
17074
|
95
|
95
|
1
|
0.00556401546210613
|
|
GO:MF
|
GO:0036094
|
small molecule binding
|
0.00175569228791398
|
2506
|
60
|
19
|
0.316666666666667
|
0.00758180367118915
|
|
GO:MF
|
GO:0003824
|
catalytic activity
|
0.00184355972834834
|
5896
|
113
|
54
|
0.47787610619469
|
0.00915875169606513
|
|
GO:MF
|
GO:0140359
|
ABC-type transporter activity
|
0.00203374702066296
|
23
|
101
|
3
|
0.0297029702970297
|
0.130434782608696
|
|
GO:MF
|
GO:0046983
|
protein dimerization activity
|
0.00225231220762138
|
1069
|
56
|
11
|
0.196428571428571
|
0.010289990645463
|
|
GO:MF
|
GO:0030545
|
receptor regulator activity
|
0.0030910638405835
|
545
|
47
|
7
|
0.148936170212766
|
0.0128440366972477
|
|
GO:MF
|
GO:0004842
|
ubiquitin-protein transferase activity
|
0.00352986760106424
|
442
|
139
|
11
|
0.079136690647482
|
0.0248868778280543
|
|
GO:MF
|
GO:0003727
|
single-stranded RNA binding
|
0.00370532546989316
|
92
|
133
|
5
|
0.037593984962406
|
0.0543478260869565
|
|
GO:MF
|
GO:0005102
|
signaling receptor binding
|
0.00400205629131674
|
1672
|
101
|
20
|
0.198019801980198
|
0.0119617224880383
|
|
GO:MF
|
GO:0019787
|
ubiquitin-like protein transferase activity
|
0.00487465283505232
|
465
|
139
|
11
|
0.079136690647482
|
0.0236559139784946
|
|
GO:MF
|
GO:0003724
|
RNA helicase activity
|
0.00608516062829325
|
76
|
106
|
4
|
0.0377358490566038
|
0.0526315789473684
|
|
GO:MF
|
GO:1904680
|
peptide transmembrane transporter activity
|
0.00680740542209865
|
37
|
101
|
3
|
0.0297029702970297
|
0.0810810810810811
|
|
GO:MF
|
GO:0019239
|
deaminase activity
|
0.00816423106578454
|
32
|
126
|
3
|
0.0238095238095238
|
0.09375
|
|
GO:MF
|
GO:0048018
|
receptor ligand activity
|
0.00838412433269387
|
494
|
81
|
8
|
0.0987654320987654
|
0.0161943319838057
|
|
GO:MF
|
GO:0016779
|
nucleotidyltransferase activity
|
0.00866042023120466
|
135
|
67
|
4
|
0.0597014925373134
|
0.0296296296296296
|
|
GO:MF
|
GO:0030546
|
signaling receptor activator activity
|
0.00874031660270563
|
501
|
81
|
8
|
0.0987654320987654
|
0.0159680638722555
|
|
GO:MF
|
GO:0048020
|
CCR chemokine receptor binding
|
0.00874031660270563
|
48
|
88
|
3
|
0.0340909090909091
|
0.0625
|
|
GO:MF
|
GO:0140098
|
catalytic activity, acting on RNA
|
0.0102585323679444
|
395
|
106
|
8
|
0.0754716981132075
|
0.020253164556962
|
|
GO:MF
|
GO:0003823
|
antigen binding
|
0.0116514652675481
|
163
|
104
|
5
|
0.0480769230769231
|
0.0306748466257669
|
|
GO:MF
|
GO:0016896
|
exoribonuclease activity, producing 5'-phosphomonoesters
|
0.0121014345115071
|
37
|
133
|
3
|
0.0225563909774436
|
0.0810810810810811
|
|
GO:MF
|
GO:0001664
|
G protein-coupled receptor binding
|
0.0130781587961221
|
292
|
88
|
6
|
0.0681818181818182
|
0.0205479452054795
|
|
GO:MF
|
GO:0004532
|
exoribonuclease activity
|
0.0132865534878716
|
39
|
133
|
3
|
0.0225563909774436
|
0.0769230769230769
|
|
GO:MF
|
GO:0003723
|
RNA binding
|
0.013651461873735
|
1943
|
113
|
22
|
0.194690265486726
|
0.011322696860525
|
|
GO:MF
|
GO:0016887
|
ATPase activity
|
0.0137168402730076
|
488
|
113
|
9
|
0.079646017699115
|
0.0184426229508197
|
|
GO:MF
|
GO:0004386
|
helicase activity
|
0.0141933453727831
|
162
|
113
|
5
|
0.0442477876106195
|
0.0308641975308642
|
|
GO:MF
|
GO:0019899
|
enzyme binding
|
0.0160409518965453
|
2074
|
95
|
20
|
0.210526315789474
|
0.00964320154291225
|
|
GO:MF
|
GO:0042887
|
amide transmembrane transporter activity
|
0.0173817791364352
|
58
|
101
|
3
|
0.0297029702970297
|
0.0517241379310345
|
|
GO:MF
|
GO:1901363
|
heterocyclic compound binding
|
0.0195680225036096
|
6228
|
69
|
34
|
0.492753623188406
|
0.0054592164418754
|
|
GO:MF
|
GO:0097159
|
organic cyclic compound binding
|
0.0229504030380716
|
6309
|
69
|
34
|
0.492753623188406
|
0.00538912664447615
|
|
GO:MF
|
GO:0042626
|
ATPase-coupled transmembrane transporter activity
|
0.0235990220627858
|
67
|
101
|
3
|
0.0297029702970297
|
0.0447761194029851
|
|
GO:MF
|
GO:0061629
|
RNA polymerase II-specific DNA-binding transcription factor binding
|
0.0239953895375565
|
281
|
77
|
5
|
0.0649350649350649
|
0.0177935943060498
|
|
GO:MF
|
GO:0043167
|
ion binding
|
0.0244163632469466
|
6057
|
57
|
28
|
0.491228070175439
|
0.00462275053656926
|
|
GO:MF
|
GO:0016740
|
transferase activity
|
0.026149180195147
|
2357
|
144
|
29
|
0.201388888888889
|
0.0123037759864234
|
|
GO:MF
|
GO:0015399
|
primary active transmembrane transporter activity
|
0.0274516853449367
|
74
|
101
|
3
|
0.0297029702970297
|
0.0405405405405405
|
|
GO:MF
|
GO:0061134
|
peptidase regulator activity
|
0.0278301349166493
|
224
|
65
|
4
|
0.0615384615384615
|
0.0178571428571429
|
|
GO:MF
|
GO:0016796
|
exonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters
|
0.0279618871304572
|
57
|
133
|
3
|
0.0225563909774436
|
0.0526315789473684
|
|
GO:MF
|
GO:0004540
|
ribonuclease activity
|
0.0288554958803367
|
111
|
133
|
4
|
0.0300751879699248
|
0.036036036036036
|
|
GO:MF
|
GO:0003714
|
transcription corepressor activity
|
0.0325282703145736
|
187
|
83
|
4
|
0.0481927710843374
|
0.0213903743315508
|
|
GO:MF
|
GO:0008270
|
zinc ion binding
|
0.0350178083005077
|
833
|
126
|
12
|
0.0952380952380952
|
0.014405762304922
|
|
GO:MF
|
GO:0030234
|
enzyme regulator activity
|
0.0368998476864681
|
1248
|
86
|
12
|
0.13953488372093
|
0.00961538461538462
|
|
GO:MF
|
GO:0016810
|
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds
|
0.0370703271687248
|
132
|
126
|
4
|
0.0317460317460317
|
0.0303030303030303
|
|
GO:MF
|
GO:0003712
|
transcription coregulator activity
|
0.0416619977342327
|
506
|
140
|
9
|
0.0642857142857143
|
0.0177865612648221
|
|
GO:MF
|
GO:0004197
|
cysteine-type endopeptidase activity
|
0.0427618163619519
|
121
|
79
|
3
|
0.0379746835443038
|
0.0247933884297521
|
|
GO:MF
|
GO:0031625
|
ubiquitin protein ligase binding
|
0.042850292291357
|
296
|
91
|
5
|
0.0549450549450549
|
0.0168918918918919
|
|
GO:MF
|
GO:0004518
|
nuclease activity
|
0.0458257613879183
|
207
|
133
|
5
|
0.037593984962406
|
0.0241545893719807
|
|
GO:MF
|
GO:0061135
|
endopeptidase regulator activity
|
0.0490816591600278
|
188
|
55
|
3
|
0.0545454545454545
|
0.0159574468085106
|
|
HP
|
HP:0030355
|
Abnormal serum interferon-gamma level
|
0.000462447294436749
|
11
|
80
|
5
|
0.0625
|
0.454545454545455
|
|
HP
|
HP:0030354
|
Abnormal serum interferon level
|
0.000462447294436749
|
11
|
80
|
5
|
0.0625
|
0.454545454545455
|
|
HP
|
HP:0011112
|
Abnormality of serum cytokine level
|
0.0177331253661078
|
23
|
80
|
5
|
0.0625
|
0.217391304347826
|
|
HP
|
HP:0011111
|
Abnormality of immune serum protein physiology
|
0.0177331253661078
|
24
|
80
|
5
|
0.0625
|
0.208333333333333
|
|
KEGG
|
KEGG:05164
|
Influenza A
|
2.14103843201096e-09
|
169
|
85
|
16
|
0.188235294117647
|
0.0946745562130177
|
|
KEGG
|
KEGG:05171
|
Coronavirus disease - COVID-19
|
1.01274556228055e-06
|
231
|
65
|
13
|
0.2
|
0.0562770562770563
|
|
KEGG
|
KEGG:05169
|
Epstein-Barr virus infection
|
1.01274556228055e-06
|
198
|
104
|
15
|
0.144230769230769
|
0.0757575757575758
|
|
KEGG
|
KEGG:05160
|
Hepatitis C
|
1.01274556228055e-06
|
157
|
63
|
11
|
0.174603174603175
|
0.0700636942675159
|
|
KEGG
|
KEGG:04621
|
NOD-like receptor signaling pathway
|
1.14526460773036e-06
|
177
|
72
|
12
|
0.166666666666667
|
0.0677966101694915
|
|
KEGG
|
KEGG:05162
|
Measles
|
2.56889601203653e-06
|
139
|
64
|
10
|
0.15625
|
0.0719424460431655
|
|
KEGG
|
KEGG:04612
|
Antigen processing and presentation
|
0.00402770981900473
|
69
|
104
|
6
|
0.0576923076923077
|
0.0869565217391304
|
|
KEGG
|
KEGG:04061
|
Viral protein interaction with cytokine and cytokine receptor
|
0.0183254975759334
|
98
|
70
|
5
|
0.0714285714285714
|
0.0510204081632653
|
|
KEGG
|
KEGG:04217
|
Necroptosis
|
0.0183254975759334
|
159
|
65
|
6
|
0.0923076923076923
|
0.0377358490566038
|
|
KEGG
|
KEGG:04062
|
Chemokine signaling pathway
|
0.0183254975759334
|
190
|
91
|
8
|
0.0879120879120879
|
0.0421052631578947
|
|
KEGG
|
KEGG:05168
|
Herpes simplex virus 1 infection
|
0.0183254975759334
|
494
|
104
|
15
|
0.144230769230769
|
0.0303643724696356
|
|
KEGG
|
KEGG:04623
|
Cytosolic DNA-sensing pathway
|
0.0354590576361164
|
62
|
84
|
4
|
0.0476190476190476
|
0.0645161290322581
|
|
KEGG
|
KEGG:05165
|
Human papillomavirus infection
|
0.0390730195898927
|
331
|
78
|
9
|
0.115384615384615
|
0.027190332326284
|
|
MIRNA
|
MIRNA:hsa-miR-146a-5p
|
hsa-miR-146a-5p
|
4.69215476207405e-13
|
200
|
67
|
16
|
0.238805970149254
|
0.08
|
|
MIRNA
|
MIRNA:hsa-miR-202-5p
|
hsa-miR-202-5p
|
0.0226772157265662
|
87
|
65
|
5
|
0.0769230769230769
|
0.0574712643678161
|
|
MIRNA
|
MIRNA:hsa-miR-1-3p
|
hsa-miR-1-3p
|
0.0432404896056282
|
915
|
90
|
16
|
0.177777777777778
|
0.0174863387978142
|
|
REAC
|
REAC:R-HSA-913531
|
Interferon Signaling
|
1.39363021145965e-42
|
191
|
121
|
43
|
0.355371900826446
|
0.225130890052356
|
|
REAC
|
REAC:R-HSA-909733
|
Interferon alpha/beta signaling
|
1.77005974442421e-36
|
66
|
115
|
28
|
0.243478260869565
|
0.424242424242424
|
|
REAC
|
REAC:R-HSA-1280215
|
Cytokine Signaling in Immune system
|
6.29996499365793e-22
|
842
|
121
|
50
|
0.413223140495868
|
0.0593824228028504
|
|
REAC
|
REAC:R-HSA-877300
|
Interferon gamma signaling
|
3.47713495940072e-16
|
87
|
121
|
18
|
0.148760330578512
|
0.206896551724138
|
|
REAC
|
REAC:R-HSA-168256
|
Immune System
|
2.01240606634078e-15
|
2139
|
122
|
67
|
0.549180327868853
|
0.0313230481533427
|
|
REAC
|
REAC:R-HSA-1169410
|
Antiviral mechanism by IFN-stimulated genes
|
6.55230181687149e-14
|
78
|
84
|
14
|
0.166666666666667
|
0.179487179487179
|
|
REAC
|
REAC:R-HSA-1169408
|
ISG15 antiviral mechanism
|
9.8347560973886e-09
|
70
|
84
|
10
|
0.119047619047619
|
0.142857142857143
|
|
REAC
|
REAC:R-HSA-8983711
|
OAS antiviral response
|
1.88018776754691e-07
|
9
|
84
|
5
|
0.0595238095238095
|
0.555555555555556
|
|
REAC
|
REAC:R-HSA-1236975
|
Antigen processing-Cross presentation
|
7.39196136029619e-07
|
99
|
121
|
11
|
0.0909090909090909
|
0.111111111111111
|
|
REAC
|
REAC:R-HSA-983169
|
Class I MHC mediated antigen processing & presentation
|
3.20224879965987e-06
|
370
|
139
|
20
|
0.143884892086331
|
0.0540540540540541
|
|
REAC
|
REAC:R-HSA-1236974
|
ER-Phagosome pathway
|
5.71756770908433e-05
|
83
|
104
|
8
|
0.0769230769230769
|
0.0963855421686747
|
|
REAC
|
REAC:R-HSA-983170
|
Antigen Presentation: Folding, assembly and peptide loading of class I MHC
|
0.000137986932737827
|
25
|
104
|
5
|
0.0480769230769231
|
0.2
|
|
REAC
|
REAC:R-HSA-936440
|
Negative regulators of DDX58/IFIH1 signaling
|
0.00021851988681211
|
34
|
84
|
5
|
0.0595238095238095
|
0.147058823529412
|
|
REAC
|
REAC:R-HSA-1280218
|
Adaptive Immune System
|
0.00125327402327709
|
800
|
139
|
25
|
0.179856115107914
|
0.03125
|
|
REAC
|
REAC:R-HSA-9683610
|
Maturation of nucleoprotein
|
0.00690662859079205
|
10
|
140
|
3
|
0.0214285714285714
|
0.3
|
|
REAC
|
REAC:R-HSA-1236978
|
Cross-presentation of soluble exogenous antigens (endosomes)
|
0.00690662859079205
|
50
|
121
|
5
|
0.0413223140495868
|
0.1
|
|
REAC
|
REAC:R-HSA-168928
|
DDX58/IFIH1-mediated induction of interferon-alpha/beta
|
0.00853913715670941
|
76
|
84
|
5
|
0.0595238095238095
|
0.0657894736842105
|
|
REAC
|
REAC:R-HSA-8939902
|
Regulation of RUNX2 expression and activity
|
0.016053640851111
|
71
|
26
|
3
|
0.115384615384615
|
0.0422535211267606
|
|
REAC
|
REAC:R-HSA-196807
|
Nicotinate metabolism
|
0.016053640851111
|
31
|
140
|
4
|
0.0285714285714286
|
0.129032258064516
|
|
REAC
|
REAC:R-HSA-983168
|
Antigen processing: Ubiquitination & Proteasome degradation
|
0.0162495560508947
|
308
|
139
|
12
|
0.0863309352517986
|
0.038961038961039
|
|
REAC
|
REAC:R-HSA-5668541
|
TNFR2 non-canonical NF-kB pathway
|
0.0371408581910769
|
100
|
26
|
3
|
0.115384615384615
|
0.03
|
|
REAC
|
REAC:R-HSA-197264
|
Nicotinamide salvaging
|
0.0371408581910769
|
19
|
140
|
3
|
0.0214285714285714
|
0.157894736842105
|
|
REAC
|
REAC:R-HSA-380108
|
Chemokine receptors bind chemokines
|
0.0383743143663414
|
57
|
47
|
3
|
0.0638297872340425
|
0.0526315789473684
|
|
TF
|
TF:M11685
|
Factor: IRF-8; motif: NCGAAACYGAAACYN
|
8.85651454265966e-41
|
1617
|
143
|
76
|
0.531468531468531
|
0.0470006184291899
|
|
TF
|
TF:M04020
|
Factor: IRF8; motif: NCGAAACCGAAACT
|
2.27855847651163e-35
|
992
|
143
|
59
|
0.412587412587413
|
0.0594758064516129
|
|
TF
|
TF:M11664
|
Factor: IRF-2; motif: NGAAASYGAAAS
|
3.40044078384031e-35
|
1651
|
143
|
71
|
0.496503496503497
|
0.0430042398546336
|
|
TF
|
TF:M09957
|
Factor: IRF-2; motif: NAAANNGAAAGTGAAASTRN
|
4.54336351221205e-35
|
272
|
143
|
38
|
0.265734265734266
|
0.139705882352941
|
|
TF
|
TF:M11665
|
Factor: IRF-2; motif: NGAAASYGAAAS
|
2.69127627008938e-34
|
2396
|
143
|
81
|
0.566433566433566
|
0.0338063439065108
|
|
TF
|
TF:M08775_1
|
Factor: IRF-2; motif: NAANYGAAASYR; match class: 1
|
1.11351802678129e-32
|
2761
|
143
|
84
|
0.587412587412587
|
0.0304237595074248
|
|
TF
|
TF:M04788
|
Factor: IRF-1; motif: STTTCACTTTCNNT
|
1.53608042308152e-32
|
536
|
143
|
45
|
0.314685314685315
|
0.083955223880597
|
|
TF
|
TF:M07216
|
Factor: IRF1; motif: NNNYASTTTCACTTTCNNTTT
|
1.8948330348184e-32
|
440
|
143
|
42
|
0.293706293706294
|
0.0954545454545455
|
|
TF
|
TF:M08887
|
Factor: IRF; motif: NNGAAANTGAAANN
|
3.05210635327109e-32
|
1267
|
143
|
61
|
0.426573426573427
|
0.0481452249408051
|
|
TF
|
TF:M11670
|
Factor: IRF-5; motif: NYGAAACCGAAACY
|
3.05210635327109e-32
|
871
|
143
|
53
|
0.370629370629371
|
0.060849598163031
|
|
TF
|
TF:M04014
|
Factor: IRF3; motif: NNRRAANGGAAACCGAAACYR
|
3.10911620427463e-32
|
278
|
143
|
36
|
0.251748251748252
|
0.129496402877698
|
|
TF
|
TF:M11679
|
Factor: IRF-7; motif: NCGAAANCGAAANYN
|
3.98247205885465e-31
|
622
|
143
|
46
|
0.321678321678322
|
0.0739549839228296
|
|
TF
|
TF:M11686
|
Factor: IRF-4; motif: NYGAAASYGAAACYN
|
9.25634851773206e-31
|
3391
|
143
|
89
|
0.622377622377622
|
0.0262459451489236
|
|
TF
|
TF:M11685_1
|
Factor: IRF-8; motif: NCGAAACYGAAACYN; match class: 1
|
1.11134281572788e-30
|
118
|
143
|
27
|
0.188811188811189
|
0.228813559322034
|
|
TF
|
TF:M09733_1
|
Factor: STAT2; motif: NRGAAANNGAAACTNA; match class: 1
|
1.334637619285e-30
|
367
|
143
|
38
|
0.265734265734266
|
0.103542234332425
|
|
TF
|
TF:M10080
|
Factor: STAT2; motif: RRGRAANNGAAACTGAAAN
|
8.4902270780548e-30
|
356
|
143
|
37
|
0.258741258741259
|
0.103932584269663
|
|
TF
|
TF:M04018
|
Factor: IRF7; motif: NCGAAARYGAAANT
|
5.31355088903251e-29
|
619
|
143
|
44
|
0.307692307692308
|
0.0710823909531502
|
|
TF
|
TF:M00453
|
Factor: IRF-7; motif: TNSGAAWNCGAAANTNNN
|
1.11663546803927e-28
|
3082
|
143
|
83
|
0.58041958041958
|
0.026930564568462
|
|
TF
|
TF:M11683_1
|
Factor: IRF-8; motif: NCGAAACCGAAACYN; match class: 1
|
2.3496539819688e-28
|
814
|
143
|
48
|
0.335664335664336
|
0.058968058968059
|
|
TF
|
TF:M09733
|
Factor: STAT2; motif: NRGAAANNGAAACTNA
|
2.61415702702876e-28
|
2842
|
144
|
80
|
0.555555555555556
|
0.0281491907107671
|
|
TF
|
TF:M04015
|
Factor: IRF-4; motif: NCGAAACCGAAACYA
|
3.2418503283396e-28
|
2038
|
143
|
69
|
0.482517482517482
|
0.0338567222767419
|
|
TF
|
TF:M11682
|
Factor: IRF-8; motif: NYGAAASYGAAACYN
|
5.10371411272053e-28
|
3795
|
143
|
90
|
0.629370629370629
|
0.0237154150197628
|
|
TF
|
TF:M11682_1
|
Factor: IRF-8; motif: NYGAAASYGAAACYN; match class: 1
|
6.20014392219531e-28
|
543
|
143
|
41
|
0.286713286713287
|
0.0755064456721915
|
|
TF
|
TF:M11664_1
|
Factor: IRF-2; motif: NGAAASYGAAAS; match class: 1
|
2.17747900835981e-27
|
140
|
140
|
26
|
0.185714285714286
|
0.185714285714286
|
|
TF
|
TF:M11687
|
Factor: IRF-4; motif: NCGAAACCGAAACYN
|
2.57121275162683e-27
|
3595
|
143
|
87
|
0.608391608391608
|
0.0242002781641168
|
|
TF
|
TF:M11671
|
Factor: IRF-5; motif: NCGAAACCGAAACY
|
2.60444045590484e-27
|
389
|
143
|
36
|
0.251748251748252
|
0.0925449871465296
|
|
TF
|
TF:M11684_1
|
Factor: IRF-8; motif: NYGAAACYGAAACTN; match class: 1
|
5.35542740926501e-27
|
832
|
143
|
47
|
0.328671328671329
|
0.0564903846153846
|
|
TF
|
TF:M11677
|
Factor: IRF-3; motif: NGGAAACNGAAACCGAAACN
|
7.82790211592367e-27
|
470
|
143
|
38
|
0.265734265734266
|
0.0808510638297872
|
|
TF
|
TF:M08887_1
|
Factor: IRF; motif: NNGAAANTGAAANN; match class: 1
|
4.53278590670551e-26
|
73
|
143
|
21
|
0.146853146853147
|
0.287671232876712
|
|
TF
|
TF:M07045
|
Factor: IRF-1; motif: NNRAAANNGAAASN
|
6.00978929640291e-26
|
535
|
143
|
39
|
0.272727272727273
|
0.0728971962616822
|
|
TF
|
TF:M11686_1
|
Factor: IRF-4; motif: NYGAAASYGAAACYN; match class: 1
|
4.38969555238635e-25
|
452
|
143
|
36
|
0.251748251748252
|
0.079646017699115
|
|
TF
|
TF:M09629
|
Factor: IRF-1; motif: NAAANNGAAAGTGAAASTRN
|
8.73842404001452e-25
|
336
|
140
|
32
|
0.228571428571429
|
0.0952380952380952
|
|
TF
|
TF:M11665_1
|
Factor: IRF-2; motif: NGAAASYGAAAS; match class: 1
|
1.11134372962849e-24
|
246
|
143
|
29
|
0.202797202797203
|
0.117886178861789
|
|
TF
|
TF:M00453_1
|
Factor: IRF-7; motif: TNSGAAWNCGAAANTNNN; match class: 1
|
1.15622543303371e-24
|
332
|
143
|
32
|
0.223776223776224
|
0.0963855421686747
|
|
TF
|
TF:M11676
|
Factor: IRF-3; motif: NGGAAANGGAAASNGAAACN
|
1.83679714402704e-24
|
562
|
131
|
37
|
0.282442748091603
|
0.0658362989323843
|
|
TF
|
TF:M00772
|
Factor: IRF; motif: RRAANTGAAASYGNV
|
4.44409413894702e-24
|
1029
|
143
|
48
|
0.335664335664336
|
0.0466472303206997
|
|
TF
|
TF:M11687_1
|
Factor: IRF-4; motif: NCGAAACCGAAACYN; match class: 1
|
2.81959295838147e-23
|
474
|
143
|
35
|
0.244755244755245
|
0.0738396624472574
|
|
TF
|
TF:M11683
|
Factor: IRF-8; motif: NCGAAACCGAAACYN
|
3.71709966293939e-23
|
4690
|
144
|
93
|
0.645833333333333
|
0.0198294243070362
|
|
TF
|
TF:M09956
|
Factor: IRF-1; motif: NAAANNGAAASTGAAASNRN
|
8.26266263111779e-23
|
317
|
143
|
30
|
0.20979020979021
|
0.0946372239747634
|
|
TF
|
TF:M11684
|
Factor: IRF-8; motif: NYGAAACYGAAACTN
|
1.34947768993029e-22
|
4664
|
144
|
92
|
0.638888888888889
|
0.0197255574614065
|
|
TF
|
TF:M00258
|
Factor: ISGF-3; motif: CAGTTTCWCTTTYCC
|
3.34953908201485e-20
|
823
|
143
|
40
|
0.27972027972028
|
0.0486026731470231
|
|
TF
|
TF:M08775
|
Factor: IRF-2; motif: NAANYGAAASYR
|
1.16682007914431e-18
|
8370
|
141
|
114
|
0.808510638297872
|
0.0136200716845878
|
|
TF
|
TF:M07323
|
Factor: IRF-4; motif: KRAAMNGAAANYN
|
3.65168794069046e-18
|
938
|
143
|
40
|
0.27972027972028
|
0.0426439232409382
|
|
TF
|
TF:M11680
|
Factor: IRF-9; motif: NYGAAACYGAAACYN
|
1.08860852761407e-17
|
128
|
143
|
19
|
0.132867132867133
|
0.1484375
|
|
TF
|
TF:M00699
|
Factor: ICSBP; motif: RAARTGAAACTG
|
4.1450245951002e-17
|
1184
|
143
|
43
|
0.300699300699301
|
0.0363175675675676
|
|
TF
|
TF:M11681
|
Factor: IRF-9; motif: NCGAAACYGAAACYN
|
5.23488077594703e-17
|
97
|
143
|
17
|
0.118881118881119
|
0.175257731958763
|
|
TF
|
TF:M04015_1
|
Factor: IRF-4; motif: NCGAAACCGAAACYA; match class: 1
|
6.01115102967599e-16
|
158
|
143
|
19
|
0.132867132867133
|
0.120253164556962
|
|
TF
|
TF:M09958
|
Factor: IRF-3; motif: RAAARGGAAANNGAAASNGA
|
6.01115102967599e-16
|
589
|
135
|
30
|
0.222222222222222
|
0.0509337860780985
|
|
TF
|
TF:M04016
|
Factor: IRF5; motif: CCGAAACCGAAACY
|
7.89641733661766e-16
|
314
|
143
|
24
|
0.167832167832168
|
0.0764331210191083
|
|
TF
|
TF:M11672
|
Factor: IRF-5; motif: NCGAAACCGAAACY
|
2.10497486579826e-15
|
120
|
143
|
17
|
0.118881118881119
|
0.141666666666667
|
|
TF
|
TF:M00972
|
Factor: IRF; motif: RAAANTGAAAN
|
1.55376172444841e-14
|
1979
|
143
|
51
|
0.356643356643357
|
0.0257705912076806
|
|
TF
|
TF:M00699_1
|
Factor: ICSBP; motif: RAARTGAAACTG; match class: 1
|
6.98144872964103e-14
|
47
|
143
|
12
|
0.0839160839160839
|
0.25531914893617
|
|
TF
|
TF:M11679_1
|
Factor: IRF-7; motif: NCGAAANCGAAANYN; match class: 1
|
1.77470916415408e-12
|
23
|
131
|
9
|
0.0687022900763359
|
0.391304347826087
|
|
TF
|
TF:M04020_1
|
Factor: IRF8; motif: NCGAAACCGAAACT; match class: 1
|
1.9620673659545e-12
|
45
|
143
|
11
|
0.0769230769230769
|
0.244444444444444
|
|
TF
|
TF:M00972_1
|
Factor: IRF; motif: RAAANTGAAAN; match class: 1
|
2.29929718100706e-12
|
205
|
72
|
14
|
0.194444444444444
|
0.0682926829268293
|
|
TF
|
TF:M11670_1
|
Factor: IRF-5; motif: NYGAAACCGAAACY; match class: 1
|
7.37767311228134e-12
|
36
|
143
|
10
|
0.0699300699300699
|
0.277777777777778
|
|
TF
|
TF:M00772_1
|
Factor: IRF; motif: RRAANTGAAASYGNV; match class: 1
|
1.36499979315621e-11
|
57
|
93
|
10
|
0.10752688172043
|
0.175438596491228
|
|
TF
|
TF:M07045_1
|
Factor: IRF-1; motif: NNRAAANNGAAASN; match class: 1
|
1.77982009695981e-11
|
19
|
126
|
8
|
0.0634920634920635
|
0.421052631578947
|
|
TF
|
TF:M10080_1
|
Factor: STAT2; motif: RRGRAANNGAAACTGAAAN; match class: 1
|
2.93304688743568e-11
|
11
|
141
|
7
|
0.049645390070922
|
0.636363636363636
|
|
TF
|
TF:M11673
|
Factor: IRF-5; motif: NCGAAACCGAAACY
|
2.98118447825035e-11
|
57
|
143
|
11
|
0.0769230769230769
|
0.192982456140351
|
|
TF
|
TF:M00258_1
|
Factor: ISGF-3; motif: CAGTTTCWCTTTYCC; match class: 1
|
5.78545133855065e-11
|
30
|
143
|
9
|
0.0629370629370629
|
0.3
|
|
TF
|
TF:M09745
|
Factor: HELIOS; motif: RNARRRGGAASTGARAN
|
1.59852399922027e-10
|
3582
|
135
|
60
|
0.444444444444444
|
0.016750418760469
|
|
TF
|
TF:M11678
|
Factor: IRF-7; motif: NNGAAAGTGAAANTR
|
1.60074112482874e-10
|
179
|
135
|
15
|
0.111111111111111
|
0.0837988826815642
|
|
TF
|
TF:M01882
|
Factor: IRF-2; motif: NNRAAAGTGAAASNNA
|
1.69821917641047e-09
|
258
|
111
|
15
|
0.135135135135135
|
0.0581395348837209
|
|
TF
|
TF:M04017
|
Factor: IRF5; motif: NACCGAAACYA
|
2.01813206410979e-09
|
195
|
123
|
14
|
0.113821138211382
|
0.0717948717948718
|
|
TF
|
TF:M04788_1
|
Factor: IRF-1; motif: STTTCACTTTCNNT; match class: 1
|
1.09740486124275e-08
|
17
|
93
|
6
|
0.0645161290322581
|
0.352941176470588
|
|
TF
|
TF:M00063
|
Factor: IRF-2; motif: GAAAAGYGAAASY
|
3.95889142369205e-08
|
685
|
138
|
23
|
0.166666666666667
|
0.0335766423357664
|
|
TF
|
TF:M11676_1
|
Factor: IRF-3; motif: NGGAAANGGAAASNGAAACN; match class: 1
|
3.97338111517076e-08
|
41
|
140
|
8
|
0.0571428571428571
|
0.195121951219512
|
|
TF
|
TF:M01066
|
Factor: Blimp-1; motif: NRGRAAGKGAAAGKRN
|
4.09125237396213e-08
|
411
|
135
|
18
|
0.133333333333333
|
0.0437956204379562
|
|
TF
|
TF:M01279
|
Factor: IRF-3; motif: NBNBTTTCSCTTT
|
2.45851790718102e-07
|
1647
|
140
|
35
|
0.25
|
0.021250758955677
|
|
TF
|
TF:M04018_1
|
Factor: IRF7; motif: NCGAAARYGAAANT; match class: 1
|
3.16433137087159e-07
|
24
|
111
|
6
|
0.0540540540540541
|
0.25
|
|
TF
|
TF:M09745_1
|
Factor: HELIOS; motif: RNARRRGGAASTGARAN; match class: 1
|
3.51958882100715e-07
|
495
|
129
|
18
|
0.13953488372093
|
0.0363636363636364
|
|
TF
|
TF:M03793
|
Factor: IRF-7; motif: AARGGAAANNGAA
|
4.17124482802541e-07
|
1987
|
143
|
39
|
0.272727272727273
|
0.019627579265224
|
|
TF
|
TF:M10040_1
|
Factor: Blimp-1; motif: RAAAGNGAAAGTNN; match class: 1
|
9.39137557627807e-07
|
12
|
135
|
5
|
0.037037037037037
|
0.416666666666667
|
|
TF
|
TF:M09957_1
|
Factor: IRF-2; motif: NAAANNGAAAGTGAAASTRN; match class: 1
|
2.72051062228143e-06
|
22
|
30
|
4
|
0.133333333333333
|
0.181818181818182
|
|
TF
|
TF:M01665_1
|
Factor: IRF-8; motif: AGTTTCW; match class: 1
|
3.09696156876331e-06
|
721
|
22
|
9
|
0.409090909090909
|
0.0124826629680999
|
|
TF
|
TF:M09629_1
|
Factor: IRF-1; motif: NAAANNGAAAGTGAAASTRN; match class: 1
|
4.0506959358776e-06
|
29
|
140
|
6
|
0.0428571428571429
|
0.206896551724138
|
|
TF
|
TF:M07216_1
|
Factor: IRF1; motif: NNNYASTTTCACTTTCNNTTT; match class: 1
|
1.31941597860743e-05
|
35
|
140
|
6
|
0.0428571428571429
|
0.171428571428571
|
|
TF
|
TF:M01066_1
|
Factor: Blimp-1; motif: NRGRAAGKGAAAGKRN; match class: 1
|
2.51732243774057e-05
|
11
|
111
|
4
|
0.036036036036036
|
0.363636363636364
|
|
TF
|
TF:M03916
|
Factor: PRDM1; motif: NRAAAGTGAAAGTNN
|
3.90523033292158e-05
|
948
|
135
|
22
|
0.162962962962963
|
0.0232067510548523
|
|
TF
|
TF:M11680_1
|
Factor: IRF-9; motif: NYGAAACYGAAACYN; match class: 1
|
4.44908259169186e-05
|
4
|
103
|
3
|
0.029126213592233
|
0.75
|
|
TF
|
TF:M07121_1
|
Factor: Blimp-1; motif: NRAAAGTGAAAGTNN; match class: 1
|
5.32205068343657e-05
|
11
|
135
|
4
|
0.0296296296296296
|
0.363636363636364
|
|
TF
|
TF:M11674
|
Factor: IRF-3; motif: GCNGTTTCCWGGAAACNGAAAC
|
9.33788220662491e-05
|
4623
|
108
|
48
|
0.444444444444444
|
0.0103828682673589
|
|
TF
|
TF:M07314_1
|
Factor: Blimp-1; motif: RGRAAGNGAAAG; match class: 1
|
0.000118387894596588
|
118
|
135
|
8
|
0.0592592592592593
|
0.0677966101694915
|
|
TF
|
TF:M01665
|
Factor: IRF-8; motif: AGTTTCW
|
0.000147645900142994
|
4771
|
48
|
27
|
0.5625
|
0.00565919094529449
|
|
TF
|
TF:M09956_1
|
Factor: IRF-1; motif: NAAANNGAAASTGAAASNRN; match class: 1
|
0.0001598472149114
|
23
|
22
|
3
|
0.136363636363636
|
0.130434782608696
|
|
TF
|
TF:M00062
|
Factor: IRF-1; motif: SAAAAGYGAAACC
|
0.000257579725381059
|
576
|
140
|
16
|
0.114285714285714
|
0.0277777777777778
|
|
TF
|
TF:M07323_1
|
Factor: IRF-4; motif: KRAAMNGAAANYN; match class: 1
|
0.000503460507440829
|
39
|
19
|
3
|
0.157894736842105
|
0.0769230769230769
|
|
TF
|
TF:M00459_1
|
Factor: STAT5B; motif: NAWTTCYNGGAAWTN; match class: 1
|
0.000643334598549552
|
1896
|
121
|
28
|
0.231404958677686
|
0.0147679324894515
|
|
TF
|
TF:M01883_1
|
Factor: IRF-4; motif: GAAARTA; match class: 1
|
0.000783036765399009
|
2948
|
65
|
24
|
0.369230769230769
|
0.00814111261872456
|
|
TF
|
TF:M03545_1
|
Factor: c-Rel; motif: NGGGAATYTCCN; match class: 1
|
0.000840674670185927
|
1011
|
53
|
12
|
0.226415094339623
|
0.0118694362017804
|
|
TF
|
TF:M03557_1
|
Factor: P50; motif: GGRRANTCCCNN; match class: 1
|
0.00101958444491075
|
1292
|
114
|
21
|
0.184210526315789
|
0.0162538699690402
|
|
TF
|
TF:M09649_1
|
Factor: Blimp-1; motif: RAAAGTGAAAGTNN; match class: 1
|
0.00141549761427196
|
10
|
111
|
3
|
0.027027027027027
|
0.3
|
|
TF
|
TF:M04016_1
|
Factor: IRF5; motif: CCGAAACCGAAACY; match class: 1
|
0.00141549761427196
|
8
|
143
|
3
|
0.020979020979021
|
0.375
|
|
TF
|
TF:M03916_1
|
Factor: PRDM1; motif: NRAAAGTGAAAGTNN; match class: 1
|
0.001588333442344
|
50
|
135
|
5
|
0.037037037037037
|
0.1
|
|
TF
|
TF:M09663
|
Factor: STAT1; motif: YCMTTTCCNGGAAAWCNCATNN
|
0.00180168379758554
|
2762
|
124
|
35
|
0.282258064516129
|
0.0126719768283852
|
|
TF
|
TF:M00346
|
Factor: GATA-1; motif: NCWGATAACA
|
0.0020475502791255
|
4376
|
73
|
32
|
0.438356164383562
|
0.00731261425959781
|
|
TF
|
TF:M00205_1
|
Factor: GR; motif: GGTACAANNTGTYCTK; match class: 1
|
0.00212664978483622
|
1482
|
54
|
14
|
0.259259259259259
|
0.00944669365721997
|
|
TF
|
TF:M11677_1
|
Factor: IRF-3; motif: NGGAAACNGAAACCGAAACN; match class: 1
|
0.00212664978483622
|
26
|
140
|
4
|
0.0285714285714286
|
0.153846153846154
|
|
TF
|
TF:M11453
|
Factor: ELFR; motif: AAMCAGGAAGTR
|
0.00219426678219963
|
488
|
117
|
12
|
0.102564102564103
|
0.0245901639344262
|
|
TF
|
TF:M01260
|
Factor: STAT1; motif: NNTTTCYNGGAARNNNNNNNNN
|
0.00225136919121029
|
2130
|
141
|
32
|
0.226950354609929
|
0.0150234741784038
|
|
TF
|
TF:M11320_1
|
Factor: C/EBPdelta; motif: NRTTGCGYAAYN; match class: 1
|
0.0022786480188021
|
2586
|
123
|
33
|
0.268292682926829
|
0.0127610208816705
|
|
TF
|
TF:M01043_1
|
Factor: CSX; motif: TSYCACTTSM; match class: 1
|
0.00250784518148072
|
1199
|
8
|
5
|
0.625
|
0.00417014178482068
|
|
TF
|
TF:M02015
|
Factor: HNF-3gamma; motif: TGTTTRYT
|
0.00251184232203192
|
2809
|
47
|
18
|
0.382978723404255
|
0.00640797436810253
|
|
TF
|
TF:M01172_1
|
Factor: PU.1; motif: NNNNYYYACTTCCTCTTTY; match class: 1
|
0.00258138617402463
|
580
|
134
|
14
|
0.104477611940299
|
0.0241379310344828
|
|
TF
|
TF:M09649
|
Factor: Blimp-1; motif: RAAAGTGAAAGTNN
|
0.00259880441769654
|
300
|
135
|
10
|
0.0740740740740741
|
0.0333333333333333
|
|
TF
|
TF:M11320
|
Factor: C/EBPdelta; motif: NRTTGCGYAAYN
|
0.00266234355008176
|
5477
|
126
|
56
|
0.444444444444444
|
0.0102245754975351
|
|
TF
|
TF:M01808_1
|
Factor: NMYC; motif: CAYCTG; match class: 1
|
0.00306229687653887
|
6149
|
56
|
32
|
0.571428571428571
|
0.00520409822735404
|
|
TF
|
TF:M09589
|
Factor: AR; motif: NGNACANNNTGTTCYNN
|
0.00306690537382424
|
5130
|
57
|
29
|
0.508771929824561
|
0.00565302144249513
|
|
TF
|
TF:M00447
|
Factor: AR; motif: AGWACATNWTGTTCT
|
0.00308665653868121
|
4183
|
40
|
20
|
0.5
|
0.0047812574707148
|
|
TF
|
TF:M07254
|
Factor: Foxc1; motif: TTVYTTTNW
|
0.00317453769943317
|
7741
|
88
|
53
|
0.602272727272727
|
0.00684666063816044
|
|
TF
|
TF:M03793_1
|
Factor: IRF-7; motif: AARGGAAANNGAA; match class: 1
|
0.00318698247220175
|
138
|
30
|
4
|
0.133333333333333
|
0.0289855072463768
|
|
TF
|
TF:M00208
|
Factor: NF-kappaB; motif: NGGGACTTTCCA
|
0.00349628355228646
|
3757
|
116
|
40
|
0.344827586206897
|
0.0106467926537131
|
|
TF
|
TF:M09933_1
|
Factor: FOXP1; motif: TNTGTTTMY; match class: 1
|
0.00450040955041636
|
634
|
57
|
9
|
0.157894736842105
|
0.0141955835962145
|
|
TF
|
TF:M00638
|
Factor: HNF4alpha; motif: VTGAACTTTGMMB
|
0.00470107159500932
|
2814
|
37
|
15
|
0.405405405405405
|
0.00533049040511727
|
|
TF
|
TF:M11674_1
|
Factor: IRF-3; motif: GCNGTTTCCWGGAAACNGAAAC; match class: 1
|
0.00471167224487176
|
1663
|
108
|
22
|
0.203703703703704
|
0.0132291040288635
|
|
TF
|
TF:M00457
|
Factor: STAT5A; motif: NAWTTCYNGGAANYN
|
0.00472913090493911
|
4038
|
121
|
43
|
0.355371900826446
|
0.0106488360574542
|
|
TF
|
TF:M09899
|
Factor: ehf; motif: NNANSAGGAAGTNNN
|
0.00481756582942964
|
457
|
117
|
11
|
0.094017094017094
|
0.0240700218818381
|
|
TF
|
TF:M00225_1
|
Factor: STAT3; motif: NGNNATTTCCSGGAARTGNNN; match class: 1
|
0.00481756582942964
|
1922
|
121
|
26
|
0.214876033057851
|
0.0135275754422477
|
|
TF
|
TF:M00641_1
|
Factor: HSF; motif: TTCCMGARGYTTC; match class: 1
|
0.00494152879496576
|
387
|
13
|
4
|
0.307692307692308
|
0.0103359173126615
|
|
TF
|
TF:M00208_1
|
Factor: NF-kappaB; motif: NGGGACTTTCCA; match class: 1
|
0.00527082506557218
|
604
|
106
|
12
|
0.113207547169811
|
0.0198675496688742
|
|
TF
|
TF:M11671_1
|
Factor: IRF-5; motif: NCGAAACCGAAACY; match class: 1
|
0.00529488836977298
|
14
|
131
|
3
|
0.0229007633587786
|
0.214285714285714
|
|
TF
|
TF:M07038_1
|
Factor: DBP; motif: TTRCATAANN; match class: 1
|
0.00536517250369465
|
772
|
84
|
12
|
0.142857142857143
|
0.0155440414507772
|
|
TF
|
TF:M04903_1
|
Factor: HNF-4alpha; motif: AGTCCAAR; match class: 1
|
0.0055282883446593
|
1534
|
72
|
16
|
0.222222222222222
|
0.0104302477183833
|
|
TF
|
TF:M00422
|
Factor: FOXJ2; motif: NNNWAAAYAAAYANNNNN
|
0.00566363766077851
|
2538
|
121
|
31
|
0.256198347107438
|
0.012214342001576
|
|
TF
|
TF:M11321
|
Factor: C/EBPdelta; motif: NRTTGCGYAAYN
|
0.00579133977971567
|
5597
|
65
|
33
|
0.507692307692308
|
0.00589601572270859
|
|
TF
|
TF:M11988_1
|
Factor: NFATc3; motif: NTTTCCRYGGAAAN; match class: 1
|
0.00579133977971567
|
6648
|
72
|
40
|
0.555555555555556
|
0.00601684717208183
|
|
TF
|
TF:M07355
|
Factor: GR; motif: AGAACAN
|
0.00624020867378998
|
5862
|
32
|
20
|
0.625
|
0.00341180484476288
|
|
TF
|
TF:M10587
|
Factor: Barx-2; motif: NWWAAYMATTAN
|
0.0062975232504715
|
1776
|
34
|
11
|
0.323529411764706
|
0.00619369369369369
|
|
TF
|
TF:M00225
|
Factor: STAT3; motif: NGNNATTTCCSGGAARTGNNN
|
0.00648801352469622
|
3188
|
121
|
36
|
0.297520661157025
|
0.0112923462986198
|
|
TF
|
TF:M10079
|
Factor: STAT1; motif: NCCNTTTCCNGGAAANCNC
|
0.00662688223003006
|
2284
|
141
|
32
|
0.226950354609929
|
0.0140105078809107
|
|
TF
|
TF:M06454
|
Factor: ZNF786; motif: KGGWTYAGGGGA
|
0.00662688223003006
|
46
|
112
|
4
|
0.0357142857142857
|
0.0869565217391304
|
|
TF
|
TF:M11571
|
Factor: HNF-3alpha; motif: YTRWGTMAATATTTRCWYWN
|
0.00684747600444945
|
4307
|
66
|
28
|
0.424242424242424
|
0.00650104481077316
|
|
TF
|
TF:M08632
|
Factor: TEF-3:Dlx-2; motif: RMATWCYNNNYAATTA
|
0.00703654983527407
|
600
|
65
|
9
|
0.138461538461538
|
0.015
|
|
TF
|
TF:M08295_1
|
Factor: FOXO1A:Elf-1; motif: NAGAAAACCGAANM; match class: 1
|
0.00703654983527407
|
4997
|
61
|
29
|
0.475409836065574
|
0.00580348208925355
|
|
TF
|
TF:M12334
|
Factor: ZBTB20; motif: NNAAYGTATRKN
|
0.00703654983527407
|
1214
|
121
|
19
|
0.15702479338843
|
0.0156507413509061
|
|
TF
|
TF:M11985_1
|
Factor: NFATc2; motif: TTTTCCATGGAAAA; match class: 1
|
0.00703654983527407
|
4518
|
105
|
41
|
0.39047619047619
|
0.00907481186365649
|
|
TF
|
TF:M11383
|
Factor: ESE-1; motif: ACCAGGAAGTANNNNNNNAAWAA
|
0.00732047110542611
|
3886
|
136
|
45
|
0.330882352941176
|
0.0115800308800823
|
|
TF
|
TF:M04317
|
Factor: EN1; motif: TAATTRSNYAATTA
|
0.00732047110542611
|
2735
|
24
|
11
|
0.458333333333333
|
0.00402193784277879
|
|
TF
|
TF:M08817
|
Factor: HOXD13; motif: WNNAATAAWAY
|
0.00771671485323447
|
5669
|
97
|
45
|
0.463917525773196
|
0.00793790792026812
|
|
TF
|
TF:M04360
|
Factor: HOXD11; motif: NNYMATAAAA
|
0.00803816469552161
|
4936
|
65
|
30
|
0.461538461538462
|
0.00607779578606159
|
|
TF
|
TF:M00155
|
Factor: ARP-1; motif: TGARCCYTTGAMCCCW
|
0.00855730987090914
|
5581
|
121
|
53
|
0.43801652892562
|
0.00949650600250851
|
|
TF
|
TF:M09958_1
|
Factor: IRF-3; motif: RAAARGGAAANNGAAASNGA; match class: 1
|
0.00862012331301274
|
93
|
112
|
5
|
0.0446428571428571
|
0.0537634408602151
|
|
TF
|
TF:M08233
|
Factor: ERF:C/EBPdelta; motif: NNCGGAWRTTGCGCAAY
|
0.00971217773607607
|
5782
|
137
|
60
|
0.437956204379562
|
0.0103770321687997
|
|
TF
|
TF:M10040
|
Factor: Blimp-1; motif: RAAAGNGAAAGTNN
|
0.00975290229356465
|
366
|
135
|
10
|
0.0740740740740741
|
0.0273224043715847
|
|
TF
|
TF:M08817_1
|
Factor: HOXD13; motif: WNNAATAAWAY; match class: 1
|
0.0102966729277217
|
1646
|
79
|
17
|
0.215189873417722
|
0.0103280680437424
|
|
TF
|
TF:M02112
|
Factor: OC-2; motif: TCAATA
|
0.0102966729277217
|
7568
|
104
|
58
|
0.557692307692308
|
0.00766384778012685
|
|
TF
|
TF:M03833
|
Factor: NKX3A; motif: NCACTTANTYN
|
0.0109652979336996
|
895
|
80
|
12
|
0.15
|
0.0134078212290503
|
|
TF
|
TF:M00223
|
Factor: STAT; motif: TTCCCGKAA
|
0.0114266393198152
|
3501
|
123
|
38
|
0.308943089430894
|
0.0108540417023708
|
|
TF
|
TF:M10586
|
Factor: Barx-2; motif: NWWAAYCATTAN
|
0.0114369044458353
|
1016
|
34
|
8
|
0.235294117647059
|
0.0078740157480315
|
|
TF
|
TF:M07204
|
Factor: AR; motif: ARGAACANNNTGTNC
|
0.0116269538474325
|
4906
|
16
|
11
|
0.6875
|
0.00224215246636771
|
|
TF
|
TF:M07229
|
Factor: STAT3; motif: NTTCYKGGAAN
|
0.0120780782283542
|
3717
|
141
|
44
|
0.312056737588652
|
0.0118375033629271
|
|
TF
|
TF:M01393
|
Factor: Msx-2; motif: NNNGACYAATTAGYNNT
|
0.0124969780795068
|
8741
|
25
|
20
|
0.8
|
0.00228806772680471
|
|
TF
|
TF:M10082
|
Factor: STAT4; motif: YTTCYYNGMAN
|
0.0125907023759407
|
882
|
133
|
16
|
0.120300751879699
|
0.018140589569161
|
|
TF
|
TF:M11378
|
Factor: ESE-1; motif: ACMAGGAAGTANNNNNNNWWAWW
|
0.0129187062940375
|
3049
|
117
|
33
|
0.282051282051282
|
0.0108232207281076
|
|
TF
|
TF:M09664
|
Factor: STAT3; motif: NTTCCYGGAAN
|
0.0129991814963635
|
5587
|
123
|
53
|
0.430894308943089
|
0.00948630749955253
|
|
TF
|
TF:M01292_1
|
Factor: HOXA13; motif: ATAAMA; match class: 1
|
0.0129991814963635
|
8366
|
75
|
47
|
0.626666666666667
|
0.00561797752808989
|
|
TF
|
TF:M03854_1
|
Factor: SRY; motif: TTGTTT; match class: 1
|
0.0129991814963635
|
9098
|
79
|
52
|
0.658227848101266
|
0.00571554187733568
|
|
TF
|
TF:M00459
|
Factor: STAT5B; motif: NAWTTCYNGGAAWTN
|
0.0129991814963635
|
3076
|
121
|
34
|
0.28099173553719
|
0.0110533159947984
|
|
TF
|
TF:M03823_1
|
Factor: FOXO1A; motif: AAACAA; match class: 1
|
0.0129991814963635
|
9098
|
79
|
52
|
0.658227848101266
|
0.00571554187733568
|
|
TF
|
TF:M08719
|
Factor: HOXB2:Sox-20; motif: ACAATRSNNNNNNNATYA
|
0.0129991814963635
|
1849
|
128
|
25
|
0.1953125
|
0.0135208220659816
|
|
TF
|
TF:M06017
|
Factor: ZNF586; motif: CCTGCMYGGTGM
|
0.0132415179269577
|
94
|
125
|
5
|
0.04
|
0.0531914893617021
|
|
TF
|
TF:M00641
|
Factor: HSF; motif: TTCCMGARGYTTC
|
0.0135237493708792
|
3599
|
81
|
28
|
0.345679012345679
|
0.00777993887190886
|
|
TF
|
TF:M11675
|
Factor: IRF-3; motif: KCNGTTTCSNSGAAACCGAAACN
|
0.0135922539835862
|
4655
|
80
|
33
|
0.4125
|
0.00708915145005371
|
|
TF
|
TF:M00190
|
Factor: C/EBP; motif: NNATTGCNNAANNN
|
0.0135922539835862
|
5586
|
28
|
17
|
0.607142857142857
|
0.00304332259219477
|
|
TF
|
TF:M09941_1
|
Factor: GR; motif: RGNACANMNTGTNCY; match class: 1
|
0.0136680379965694
|
2046
|
55
|
15
|
0.272727272727273
|
0.00733137829912024
|
|
TF
|
TF:M08216
|
Factor: Elk-1:FOXI1; motif: ACCGGAAGTRTMAACAN
|
0.0136680379965694
|
7126
|
62
|
36
|
0.580645161290323
|
0.00505192253718776
|
|
TF
|
TF:M07316
|
Factor: C/EBPdelta; motif: TKNNGCAATN
|
0.0137104944547705
|
3306
|
136
|
39
|
0.286764705882353
|
0.0117967332123412
|
|
TF
|
TF:M07314
|
Factor: Blimp-1; motif: RGRAAGNGAAAG
|
0.0142531389953218
|
1460
|
135
|
22
|
0.162962962962963
|
0.0150684931506849
|
|
TF
|
TF:M04201
|
Factor: CEBPB; motif: ATTRCGCAAY
|
0.0148252075764581
|
4048
|
19
|
11
|
0.578947368421053
|
0.00271739130434783
|
|
TF
|
TF:M10170
|
Factor: ZNF85; motif: ANAGAARWCTGMWGKAATCT
|
0.0148252075764581
|
181
|
16
|
3
|
0.1875
|
0.0165745856353591
|
|
TF
|
TF:M00457_1
|
Factor: STAT5A; motif: NAWTTCYNGGAANYN; match class: 1
|
0.0151705291514522
|
1589
|
117
|
21
|
0.179487179487179
|
0.013215859030837
|
|
TF
|
TF:M00747_1
|
Factor: IRF-1; motif: TTCACTT; match class: 1
|
0.0159093024906788
|
933
|
22
|
6
|
0.272727272727273
|
0.00643086816720257
|
|
TF
|
TF:M09882
|
Factor: C/EBPdelta; motif: NTTGCNCMAYN
|
0.0159093024906788
|
2027
|
140
|
28
|
0.2
|
0.0138135175135668
|
|
TF
|
TF:M05445_1
|
Factor: GATA-5; motif: TTAMCAR; match class: 1
|
0.0159093024906788
|
414
|
47
|
6
|
0.127659574468085
|
0.0144927536231884
|
|
TF
|
TF:M07419
|
Factor: FOXP3; motif: NNNVAAACANWD
|
0.0160872145064882
|
6311
|
79
|
40
|
0.506329113924051
|
0.00633813975598162
|
|
TF
|
TF:M08413
|
Factor: TEF-3:C/EBPdelta; motif: RGWATGYNRTTRCGYAAY
|
0.0161428789737268
|
6035
|
72
|
36
|
0.5
|
0.00596520298260149
|
|
TF
|
TF:M03564_1
|
Factor: SATB1; motif: NTTTAT; match class: 1
|
0.0167217446548192
|
12003
|
144
|
107
|
0.743055555555556
|
0.00891443805715238
|
|
TF
|
TF:M01279_1
|
Factor: IRF-3; motif: NBNBTTTCSCTTT; match class: 1
|
0.0172873112305635
|
101
|
30
|
3
|
0.1
|
0.0297029702970297
|
|
TF
|
TF:M09596
|
Factor: C/EBPalpha; motif: NRTTGTGCAAYNN
|
0.0174487885436673
|
5213
|
136
|
54
|
0.397058823529412
|
0.010358718588145
|
|
TF
|
TF:M07298
|
Factor: Msx-1; motif: AHAWWTSMYYCAATTAN
|
0.0174942509960281
|
1938
|
72
|
17
|
0.236111111111111
|
0.0087719298245614
|
|
TF
|
TF:M04715_1
|
Factor: Oct-2; motif: ATTTGCA; match class: 1
|
0.0175592526302455
|
1767
|
100
|
20
|
0.2
|
0.0113186191284663
|
|
TF
|
TF:M10079_1
|
Factor: STAT1; motif: NCCNTTTCCNGGAAANCNC; match class: 1
|
0.0178208007372732
|
1164
|
117
|
17
|
0.145299145299145
|
0.0146048109965636
|
|
TF
|
TF:M02117
|
Factor: STAT4; motif: TTMNNRGAAA
|
0.0179670560827418
|
1353
|
139
|
21
|
0.151079136690647
|
0.0155210643015521
|
|
TF
|
TF:M09929
|
Factor: FOXK1; motif: TGTTTNCNNT
|
0.0179670560827418
|
4139
|
105
|
37
|
0.352380952380952
|
0.00893935733268906
|
|
TF
|
TF:M02096
|
Factor: ipf1; motif: YTAATGN
|
0.0179670560827418
|
2033
|
5
|
4
|
0.8
|
0.00196753566158387
|
|
TF
|
TF:M00791
|
Factor: HNF3; motif: NWRARYAAAYANN
|
0.0179963342952009
|
2732
|
23
|
10
|
0.434782608695652
|
0.00366032210834553
|
|
TF
|
TF:M04431
|
Factor: PRRX1; motif: NYAATTAN
|
0.0180636136054346
|
3464
|
90
|
29
|
0.322222222222222
|
0.00837182448036951
|
|
TF
|
TF:M00687
|
Factor: alpha-CP1; motif: CAGCCAATGAG
|
0.0184537641366734
|
2326
|
18
|
8
|
0.444444444444444
|
0.00343938091143594
|
|
TF
|
TF:M11321_1
|
Factor: C/EBPdelta; motif: NRTTGCGYAAYN; match class: 1
|
0.0184537641366734
|
1852
|
125
|
24
|
0.192
|
0.0129589632829374
|
|
TF
|
TF:M07228
|
Factor: STAT1; motif: NTTCYRGGAAA
|
0.0185892523371694
|
1919
|
121
|
24
|
0.198347107438017
|
0.0125065138092757
|
|
TF
|
TF:M01797
|
Factor: SIRT6; motif: AGATAARN
|
0.0185892523371694
|
1200
|
25
|
7
|
0.28
|
0.00583333333333333
|
|
TF
|
TF:M07988_1
|
Factor: EMX1; motif: NNTAATKANN; match class: 1
|
0.0185892523371694
|
1621
|
86
|
17
|
0.197674418604651
|
0.0104873534855028
|
|
TF
|
TF:M00194_1
|
Factor: NF-kappaB; motif: NGGGGAMTTTCCNN; match class: 1
|
0.0187865989207555
|
385
|
53
|
6
|
0.113207547169811
|
0.0155844155844156
|
|
TF
|
TF:M04681
|
Factor: JunD; motif: RTGACGTCA
|
0.0189224286342549
|
6003
|
115
|
52
|
0.452173913043478
|
0.00866233549891721
|
|
TF
|
TF:M11286
|
Factor: Fra-1; motif: NATGAWTCAYN
|
0.0189224286342549
|
3150
|
72
|
23
|
0.319444444444444
|
0.0073015873015873
|
|
TF
|
TF:M08359_1
|
Factor: HOXD12:Elk-1; motif: NRNCGGAAGTCGTAAAN; match class: 1
|
0.0189836845293064
|
1166
|
68
|
12
|
0.176470588235294
|
0.0102915951972556
|
|
TF
|
TF:M00054_1
|
Factor: NF-kappaB; motif: GGGAMTTYCC; match class: 1
|
0.0189967828628772
|
712
|
53
|
8
|
0.150943396226415
|
0.0112359550561798
|
|
TF
|
TF:M04324
|
Factor: GBX2; motif: NNYAATTANN
|
0.0189967828628772
|
3485
|
90
|
29
|
0.322222222222222
|
0.00832137733142037
|
|
TF
|
TF:M03865
|
Factor: Blimp-1; motif: NRGRAAGKGAAAGK
|
0.0194575779872133
|
1048
|
21
|
6
|
0.285714285714286
|
0.00572519083969466
|
|
TF
|
TF:M02079
|
Factor: Isl2; motif: CTAATKR
|
0.0194575779872133
|
5734
|
6
|
6
|
1
|
0.00104638995465644
|
|
TF
|
TF:M11589
|
Factor: FOXK1; motif: NNNGTAAACAN
|
0.0196026778094594
|
4486
|
59
|
25
|
0.423728813559322
|
0.00557289344627731
|
|
TF
|
TF:M04325
|
Factor: GSC2; motif: NNTAATCCNN
|
0.0199945812020852
|
3592
|
42
|
17
|
0.404761904761905
|
0.00473273942093541
|
|
TF
|
TF:M01996
|
Factor: AR; motif: GNNCNNNNTGTTCTN
|
0.0200742703647413
|
3085
|
36
|
14
|
0.388888888888889
|
0.00453808752025932
|
|
TF
|
TF:M10690
|
Factor: HOXA7; motif: GYMATTAN
|
0.0209254847698118
|
10748
|
55
|
42
|
0.763636363636364
|
0.00390770375883885
|
|
TF
|
TF:M12067
|
Factor: ZNF32; motif: NYGTAACNYGAYACC
|
0.0212107518621837
|
164
|
84
|
5
|
0.0595238095238095
|
0.0304878048780488
|
|
TF
|
TF:M11316
|
Factor: C/EBPbeta; motif: NRTTGCGYAAYN
|
0.0212107518621837
|
5225
|
21
|
13
|
0.619047619047619
|
0.00248803827751196
|
|
TF
|
TF:M11988
|
Factor: NFATc3; motif: NTTTCCRYGGAAAN
|
0.0212107518621837
|
7175
|
72
|
40
|
0.555555555555556
|
0.00557491289198606
|
|
TF
|
TF:M10941
|
Factor: VSX1; motif: CYAATTRN
|
0.0212107518621837
|
3970
|
103
|
35
|
0.339805825242718
|
0.00881612090680101
|
|
TF
|
TF:M00809
|
Factor: FOX; motif: KWTTGTTTRTTTW
|
0.0212107518621837
|
3746
|
137
|
42
|
0.306569343065693
|
0.0112119594233849
|
|
TF
|
TF:M11566
|
Factor: HNF-3alpha; motif: NWRWGTMAAYAW
|
0.0212107518621837
|
3516
|
62
|
22
|
0.354838709677419
|
0.00625711035267349
|
|
TF
|
TF:M00070
|
Factor: Tal-1beta:ITF-2; motif: NNNAACAGATGKTNNN
|
0.0213110826565886
|
1763
|
67
|
15
|
0.223880597014925
|
0.00850822461712989
|
|
TF
|
TF:M00465
|
Factor: POU6F1; motif: GCATAAWTTAT
|
0.0214350656616938
|
1511
|
62
|
13
|
0.209677419354839
|
0.0086035737921906
|
|
TF
|
TF:M00130
|
Factor: FOXD3; motif: NAWTGTTTRTTT
|
0.0214350656616938
|
4211
|
138
|
46
|
0.333333333333333
|
0.010923771075754
|
|
TF
|
TF:M01886
|
Factor: NFATc3; motif: GGAAAA
|
0.0214350656616938
|
16226
|
89
|
84
|
0.943820224719101
|
0.00517687661777394
|
|
TF
|
TF:M02108_1
|
Factor: CSX; motif: NNCACTTGNRN; match class: 1
|
0.0214350656616938
|
1533
|
135
|
22
|
0.162962962962963
|
0.0143509458577952
|
|
TF
|
TF:M03555
|
Factor: NFATc2; motif: GGAAAA
|
0.0214350656616938
|
16226
|
89
|
84
|
0.943820224719101
|
0.00517687661777394
|
|
TF
|
TF:M01281
|
Factor: NFATc2; motif: GGAAAA
|
0.0214350656616938
|
16226
|
89
|
84
|
0.943820224719101
|
0.00517687661777394
|
|
TF
|
TF:M09902
|
Factor: ESE-1; motif: NNANVAGGAAGTNN
|
0.0217328565041982
|
3200
|
117
|
33
|
0.282051282051282
|
0.0103125
|
|
TF
|
TF:M11317
|
Factor: C/EBPbeta; motif: NRTTGCGYAAYN
|
0.0217328565041982
|
4365
|
71
|
28
|
0.394366197183099
|
0.00641466208476518
|
|
TF
|
TF:M01230_1
|
Factor: ZNF333; motif: ATAAT; match class: 1
|
0.0217328565041982
|
11582
|
77
|
59
|
0.766233766233766
|
0.00509411155240891
|
|
TF
|
TF:M01181
|
Factor: Nkx3-2; motif: TRAGTG
|
0.0217328565041982
|
9457
|
105
|
67
|
0.638095238095238
|
0.00708469916463995
|
|
TF
|
TF:M09589_1
|
Factor: AR; motif: NGNACANNNTGTTCYNN; match class: 1
|
0.0222253776634789
|
1159
|
20
|
6
|
0.3
|
0.00517687661777394
|
|
TF
|
TF:M00476
|
Factor: FOXO4; motif: NNGTTGTTTACNTN
|
0.0223768611548728
|
2740
|
64
|
19
|
0.296875
|
0.00693430656934307
|
|
TF
|
TF:M10526_1
|
Factor: Blimp-1; motif: RRGNGAAAGNRANNN; match class: 1
|
0.0223768611548728
|
286
|
135
|
8
|
0.0592592592592593
|
0.027972027972028
|
|
TF
|
TF:M03563
|
Factor: RelA-p65; motif: GGGANTTTCCNN
|
0.0223768611548728
|
4643
|
55
|
24
|
0.436363636363636
|
0.00516907172087013
|
|
TF
|
TF:M11847_1
|
Factor: GR; motif: RGWACATWATGTWCY; match class: 1
|
0.0223768611548728
|
4066
|
55
|
22
|
0.4
|
0.00541072306935563
|
|
TF
|
TF:M03882
|
Factor: RelB:p50; motif: RGAAANTCCCYNNHGC
|
0.0223768611548728
|
4474
|
60
|
25
|
0.416666666666667
|
0.00558784085829236
|
|
TF
|
TF:M02014
|
Factor: HNF-3beta; motif: NTRTTTRYT
|
0.0223768611548728
|
2650
|
61
|
18
|
0.295081967213115
|
0.00679245283018868
|
|
TF
|
TF:M08004
|
Factor: SHOX; motif: NTAATTRN
|
0.0224979429621002
|
2226
|
66
|
17
|
0.257575757575758
|
0.00763701707097934
|
|
TF
|
TF:M00774
|
Factor: NF-kappaB; motif: NGGGANTTYCCMNNNN
|
0.0227680233977549
|
1818
|
53
|
13
|
0.245283018867925
|
0.00715071507150715
|
|
TF
|
TF:M10541_1
|
Factor: SUHW1; motif: TCTCTCCAGTRTGAATTCTCTGAT; match class: 1
|
0.0227692644961682
|
1794
|
67
|
15
|
0.223880597014925
|
0.00836120401337793
|
|
TF
|
TF:M00621
|
Factor: C/EBPdelta; motif: MATTKCNTMAYY
|
0.0231693309658395
|
3888
|
72
|
26
|
0.361111111111111
|
0.00668724279835391
|
|
TF
|
TF:M11458
|
Factor: nerf; motif: AANCMGGAAGTR
|
0.0231693309658395
|
505
|
117
|
10
|
0.0854700854700855
|
0.0198019801980198
|
|
TF
|
TF:M04745_1
|
Factor: CTCF; motif: CANNWRRTGGCAG; match class: 1
|
0.0236654936917662
|
2298
|
8
|
5
|
0.625
|
0.00217580504786771
|
|
TF
|
TF:M11445
|
Factor: Elf-1; motif: NANCAGGAAGTR
|
0.0237655255044556
|
1689
|
117
|
21
|
0.179487179487179
|
0.0124333925399645
|
|
TF
|
TF:M02015_1
|
Factor: HNF-3gamma; motif: TGTTTRYT; match class: 1
|
0.0242980984666592
|
742
|
31
|
6
|
0.193548387096774
|
0.00808625336927224
|
|
TF
|
TF:M09902_1
|
Factor: ESE-1; motif: NNANVAGGAAGTNN; match class: 1
|
0.0244158458474143
|
306
|
129
|
8
|
0.062015503875969
|
0.0261437908496732
|
|
TF
|
TF:M08520_1
|
Factor: CDP:SRF; motif: NCCWTAYAAGGTMNKRATCRATN; match class: 1
|
0.0250862418924995
|
3278
|
47
|
17
|
0.361702127659574
|
0.00518608907870653
|
|
TF
|
TF:M01866
|
Factor: C/EBPalpha; motif: NNNTTNNGCAANN
|
0.0250862418924995
|
5089
|
27
|
15
|
0.555555555555556
|
0.00294753389663981
|
|
TF
|
TF:M11985
|
Factor: NFATc2; motif: TTTTCCATGGAAAA
|
0.0257705834467219
|
5845
|
74
|
35
|
0.472972972972973
|
0.00598802395209581
|
|
TF
|
TF:M09597_1
|
Factor: C/EBPbeta; motif: NATTGCRYAAYN; match class: 1
|
0.0257705834467219
|
374
|
24
|
4
|
0.166666666666667
|
0.0106951871657754
|
|
TF
|
TF:M03547
|
Factor: ER-alpha; motif: TGACCYN
|
0.0257705834467219
|
4712
|
9
|
7
|
0.777777777777778
|
0.00148556876061121
|
|
TF
|
TF:M10003
|
Factor: NF-kappaB1; motif: AGGGAAAK
|
0.0257705834467219
|
1983
|
122
|
24
|
0.19672131147541
|
0.0121028744326778
|
|
TF
|
TF:M04434
|
Factor: RAX2; motif: NYAATTAN
|
0.0270120820103864
|
2944
|
75
|
22
|
0.293333333333333
|
0.00747282608695652
|
|
TF
|
TF:M04855_1
|
Factor: IRF-4; motif: AAGTTTC; match class: 1
|
0.0270120820103864
|
2009
|
121
|
24
|
0.198347107438017
|
0.0119462419113987
|
|
TF
|
TF:M11559
|
Factor: FKHL14; motif: NYAMAYAAACAN
|
0.0272485019161087
|
2701
|
47
|
15
|
0.319148936170213
|
0.00555349870418364
|
|
TF
|
TF:M04514_1
|
Factor: CPEB1; motif: AATAAAAA; match class: 1
|
0.0274322750756591
|
1113
|
66
|
11
|
0.166666666666667
|
0.00988319856244385
|
|
TF
|
TF:M00339_1
|
Factor: c-Ets-1; motif: RCAGGAAGTGNNTNS; match class: 1
|
0.0275185208081249
|
1466
|
51
|
11
|
0.215686274509804
|
0.00750341064120055
|
|
TF
|
TF:M11626_1
|
Factor: Sox-9; motif: NNNACAATRG; match class: 1
|
0.0275185208081249
|
1612
|
61
|
13
|
0.213114754098361
|
0.00806451612903226
|
|
TF
|
TF:M11570
|
Factor: HNF-3alpha; motif: CNYTAAGTAAACAAAN
|
0.0275185208081249
|
3651
|
6
|
5
|
0.833333333333333
|
0.00136948781155848
|
|
TF
|
TF:M07080
|
Factor: C/EBPbeta; motif: KATTGCAYMAY
|
0.0275185208081249
|
2026
|
100
|
21
|
0.21
|
0.010365251727542
|
|
TF
|
TF:M04433
|
Factor: PRRX2; motif: NYAATTAN
|
0.0275185208081249
|
3153
|
75
|
23
|
0.306666666666667
|
0.00729464002537266
|
|
TF
|
TF:M00912
|
Factor: C/EBP; motif: NTTRCNNAANNN
|
0.0276190560937523
|
4968
|
140
|
52
|
0.371428571428571
|
0.0104669887278583
|
|
TF
|
TF:M10924
|
Factor: Gscl; motif: NTAATCCN
|
0.0276190560937523
|
6815
|
79
|
41
|
0.518987341772152
|
0.00601614086573734
|
|
TF
|
TF:M04202
|
Factor: CEBPD; motif: RTTRCGCAAY
|
0.0277448810446533
|
5031
|
20
|
12
|
0.6
|
0.00238521168753727
|
|
TF
|
TF:M07254_1
|
Factor: Foxc1; motif: TTVYTTTNW; match class: 1
|
0.0294012847266042
|
2610
|
22
|
9
|
0.409090909090909
|
0.00344827586206897
|
|
TF
|
TF:M00006
|
Factor: MEF-2A; motif: CTCTAAAAATAACYCY
|
0.029612225355674
|
1986
|
18
|
7
|
0.388888888888889
|
0.00352467270896274
|
|
TF
|
TF:M07466_1
|
Factor: Six-1; motif: CTCARRTTWCN; match class: 1
|
0.0297200485481704
|
413
|
23
|
4
|
0.173913043478261
|
0.00968523002421308
|
|
TF
|
TF:M11248
|
Factor: Luman; motif: GRTGAYGTCAYN
|
0.0297200485481704
|
305
|
52
|
5
|
0.0961538461538462
|
0.0163934426229508
|
|
TF
|
TF:M04354_1
|
Factor: HOXC11; motif: NGYMATAAAAN; match class: 1
|
0.0297640174274068
|
339
|
47
|
5
|
0.106382978723404
|
0.0147492625368732
|
|
TF
|
TF:M04202_1
|
Factor: CEBPD; motif: RTTRCGCAAY; match class: 1
|
0.0297640174274068
|
1980
|
110
|
22
|
0.2
|
0.0111111111111111
|
|
TF
|
TF:M04306
|
Factor: DPRX; motif: NNGGATTANN
|
0.0300452373011794
|
4886
|
60
|
26
|
0.433333333333333
|
0.00532132623823168
|
|
TF
|
TF:M09911_1
|
Factor: c-Ets-1; motif: NNRCMGGAAGTGG; match class: 1
|
0.0303491784191914
|
208
|
45
|
4
|
0.0888888888888889
|
0.0192307692307692
|
|
TF
|
TF:M01836
|
Factor: GR; motif: CNNNNTGTYCTNN
|
0.0304406221913216
|
5942
|
36
|
20
|
0.555555555555556
|
0.00336587007741501
|
|
TF
|
TF:M09959
|
Factor: IRF-4; motif: NAANGRGGAASTGAAASN
|
0.0304406221913216
|
3386
|
28
|
12
|
0.428571428571429
|
0.00354400472533963
|
|
TF
|
TF:M11697_1
|
Factor: MEF-2B1; motif: CCNWATWNRG; match class: 1
|
0.0304406221913216
|
683
|
7
|
3
|
0.428571428571429
|
0.00439238653001464
|
|
TF
|
TF:M06132
|
Factor: ZNF823; motif: NYYACCAGCCCT
|
0.0304406221913216
|
55
|
77
|
3
|
0.038961038961039
|
0.0545454545454545
|
|
TF
|
TF:M03557
|
Factor: P50; motif: GGRRANTCCCNN
|
0.0304406221913216
|
4967
|
115
|
44
|
0.382608695652174
|
0.0088584658747735
|
|
TF
|
TF:M00291_1
|
Factor: Freac-3; motif: NNNNNGTAAATAAACA; match class: 1
|
0.0309181280426376
|
1096
|
79
|
12
|
0.151898734177215
|
0.0109489051094891
|
|
TF
|
TF:M04236
|
Factor: FOXC2; motif: RTAAAYAAACA
|
0.0310306336112544
|
576
|
128
|
11
|
0.0859375
|
0.0190972222222222
|
|
TF
|
TF:M08202
|
Factor: CDP:HOXB13; motif: SYMRTAAANATYGATY
|
0.0311076437400219
|
4079
|
78
|
28
|
0.358974358974359
|
0.00686442755577347
|
|
TF
|
TF:M10046_1
|
Factor: c-Rel; motif: NRNRGGGRAATKCCA; match class: 1
|
0.0311459733434412
|
734
|
72
|
9
|
0.125
|
0.0122615803814714
|
|
TF
|
TF:M01281_1
|
Factor: NFATc2; motif: GGAAAA; match class: 1
|
0.0311459733434412
|
11109
|
73
|
54
|
0.73972602739726
|
0.00486092357547934
|
|
TF
|
TF:M01886_1
|
Factor: NFATc3; motif: GGAAAA; match class: 1
|
0.0311459733434412
|
11109
|
73
|
54
|
0.73972602739726
|
0.00486092357547934
|
|
TF
|
TF:M03555_1
|
Factor: NFATc2; motif: GGAAAA; match class: 1
|
0.0311459733434412
|
11109
|
73
|
54
|
0.73972602739726
|
0.00486092357547934
|
|
TF
|
TF:M10963
|
Factor: RHOXF1; motif: NGGATCAN
|
0.0311459733434412
|
2641
|
128
|
30
|
0.234375
|
0.011359333585763
|
|
TF
|
TF:M04619
|
Factor: MTF-1; motif: CCGNGTGCAV
|
0.0314743533750326
|
1167
|
22
|
6
|
0.272727272727273
|
0.0051413881748072
|
|
TF
|
TF:M11727
|
Factor: NF-1B; motif: NTTGGCNNNNTGCCARN
|
0.0316158342683107
|
75
|
58
|
3
|
0.0517241379310345
|
0.04
|
|
TF
|
TF:M08469
|
Factor: Fli-1:C/EBPdelta; motif: NNCGGAWATTGCGCAAT
|
0.0316388018925053
|
6743
|
144
|
67
|
0.465277777777778
|
0.00993623016461516
|
|
TF
|
TF:M11588
|
Factor: FOXK1; motif: NWYGTAAAYAR
|
0.0316388018925053
|
5352
|
53
|
25
|
0.471698113207547
|
0.0046711509715994
|
|
TF
|
TF:M01211
|
Factor: PARP; motif: YDRGAAAWAS
|
0.0316430018240395
|
2634
|
5
|
4
|
0.8
|
0.00151860288534548
|
|
TF
|
TF:M09930_1
|
Factor: foxm1; motif: TGTTTRCTYWNN; match class: 1
|
0.0317778245781628
|
1545
|
23
|
7
|
0.304347826086957
|
0.00453074433656958
|
|
TF
|
TF:M01107
|
Factor: RUSH-1alpha; motif: NNMCWTNKNN
|
0.0322790856977274
|
3108
|
23
|
10
|
0.434782608695652
|
0.00321750321750322
|
|
TF
|
TF:M08805
|
Factor: EAR2; motif: NCTYTGACCTN
|
0.0323581591367301
|
1027
|
25
|
6
|
0.24
|
0.00584225900681597
|
|
TF
|
TF:M09625
|
Factor: GR; motif: RGNACANKNTGTNCY
|
0.0323581591367301
|
9274
|
58
|
39
|
0.672413793103448
|
0.00420530515419452
|
|
TF
|
TF:M00054
|
Factor: NF-kappaB; motif: GGGAMTTYCC
|
0.0323581591367301
|
2444
|
53
|
15
|
0.283018867924528
|
0.00613747954173486
|
|
TF
|
TF:M01408
|
Factor: Brn-3c; motif: NNTTATTAATKANKNC
|
0.0323581591367301
|
4888
|
98
|
38
|
0.387755102040816
|
0.00777414075286416
|
|
TF
|
TF:M11286_1
|
Factor: Fra-1; motif: NATGAWTCAYN; match class: 1
|
0.0324479761973902
|
2340
|
72
|
18
|
0.25
|
0.00769230769230769
|
|
TF
|
TF:M03816_1
|
Factor: c-MAF; motif: CNNNCTCAGCA; match class: 1
|
0.0324479761973902
|
1059
|
129
|
16
|
0.124031007751938
|
0.0151085930122757
|
|
TF
|
TF:M00023
|
Factor: HOXA5; motif: TGCNHNCWYCCYCATTAKTGNDCNMNHYCN
|
0.0324479761973902
|
3491
|
134
|
38
|
0.283582089552239
|
0.0108851331996563
|
|
TF
|
TF:M10818_1
|
Factor: HOXC11; motif: NGYMATAAAAN; match class: 1
|
0.0324479761973902
|
95
|
47
|
3
|
0.0638297872340425
|
0.0315789473684211
|
|
TF
|
TF:M08899
|
Factor: SOX; motif: NNNNCWTTGTT
|
0.0324479761973902
|
2066
|
6
|
4
|
0.666666666666667
|
0.00193610842207164
|
|
TF
|
TF:M01224
|
Factor: P50:RELA-P65; motif: GGANTTYCCCWN
|
0.0328088033117937
|
2329
|
139
|
29
|
0.20863309352518
|
0.0124516960068699
|
|
TF
|
TF:M07988
|
Factor: EMX1; motif: NNTAATKANN
|
0.0329846520565743
|
3512
|
103
|
31
|
0.300970873786408
|
0.00882687927107061
|
|
TF
|
TF:M09855_1
|
Factor: FOXP1; motif: CTGTTTNYTYTKN; match class: 1
|
0.0337662370363425
|
2166
|
103
|
22
|
0.213592233009709
|
0.0101569713758079
|
|
TF
|
TF:M00953
|
Factor: AR; motif: NNNGNRRGNACANNGTGTTCTNNNNNN
|
0.0337662370363425
|
2415
|
54
|
15
|
0.277777777777778
|
0.0062111801242236
|
|
TF
|
TF:M04279_1
|
Factor: ALX3; motif: NNYAATTANN; match class: 1
|
0.0337662370363425
|
1952
|
79
|
17
|
0.215189873417722
|
0.00870901639344262
|
|
TF
|
TF:M07417_1
|
Factor: Elf-1; motif: NNCAGGAAGNNN; match class: 1
|
0.0337813367082345
|
102
|
45
|
3
|
0.0666666666666667
|
0.0294117647058824
|
|
TF
|
TF:M09855
|
Factor: FOXP1; motif: CTGTTTNYTYTKN
|
0.0346862166781403
|
7645
|
105
|
56
|
0.533333333333333
|
0.00732504905166776
|
|
TF
|
TF:M07429
|
Factor: SMAD3; motif: CAGACAS
|
0.035142612125129
|
6273
|
26
|
16
|
0.615384615384615
|
0.00255061374143153
|
|
TF
|
TF:M11363
|
Factor: c-MAF; motif: NTGCTGACNNWGCA
|
0.0358210374891163
|
422
|
81
|
7
|
0.0864197530864197
|
0.0165876777251185
|
|
TF
|
TF:M00774_1
|
Factor: NF-kappaB; motif: NGGGANTTYCCMNNNN; match class: 1
|
0.0359026527553053
|
192
|
53
|
4
|
0.0754716981132075
|
0.0208333333333333
|
|
TF
|
TF:M07375
|
Factor: BCL-6; motif: TTCCTAGAAA
|
0.0359026527553053
|
556
|
117
|
10
|
0.0854700854700855
|
0.0179856115107914
|
|
TF
|
TF:M00348
|
Factor: GATA-2; motif: NNWGATAASA
|
0.0360544236310184
|
2155
|
6
|
4
|
0.666666666666667
|
0.00185614849187935
|
|
TF
|
TF:M00289_1
|
Factor: HFH3; motif: KNNTRTTTRTTTA; match class: 1
|
0.0360544236310184
|
817
|
43
|
7
|
0.162790697674419
|
0.00856793145654835
|
|
TF
|
TF:M00065
|
Factor: Tal-1beta:E47; motif: NNNAACAGATGKTNNN
|
0.0360544236310184
|
1384
|
74
|
13
|
0.175675675675676
|
0.00939306358381503
|
|
TF
|
TF:M07121
|
Factor: Blimp-1; motif: NRAAAGTGAAAGTNN
|
0.0360544236310184
|
401
|
135
|
9
|
0.0666666666666667
|
0.0224438902743142
|
|
TF
|
TF:M08295
|
Factor: FOXO1A:Elf-1; motif: NAGAAAACCGAANM
|
0.0362113896519055
|
11711
|
53
|
42
|
0.792452830188679
|
0.00358637178720861
|
|
TF
|
TF:M00195
|
Factor: Oct-1; motif: NNNNATGCAAATNAN
|
0.036305931485634
|
3659
|
121
|
36
|
0.297520661157025
|
0.00983875375785734
|
|
TF
|
TF:M01107_1
|
Factor: RUSH-1alpha; motif: NNMCWTNKNN; match class: 1
|
0.036305931485634
|
385
|
13
|
3
|
0.230769230769231
|
0.00779220779220779
|
|
TF
|
TF:M11387
|
Factor: elf5; motif: NNNNMGGAAGTN
|
0.036305931485634
|
2598
|
104
|
25
|
0.240384615384615
|
0.00962278675904542
|
|
TF
|
TF:M01260_1
|
Factor: STAT1; motif: NNTTTCYNGGAARNNNNNNNNN; match class: 1
|
0.036305931485634
|
657
|
117
|
11
|
0.094017094017094
|
0.0167427701674277
|
|
TF
|
TF:M00133_1
|
Factor: Tst-1; motif: NNKGAATTAVAVTDN; match class: 1
|
0.036305931485634
|
1298
|
21
|
6
|
0.285714285714286
|
0.00462249614791988
|
|
TF
|
TF:M00192
|
Factor: GR; motif: NNNNNNCNNTNTGTNCTNN
|
0.036305931485634
|
7611
|
36
|
23
|
0.638888888888889
|
0.00302194192615951
|
|
TF
|
TF:M00794
|
Factor: TTF-1; motif: NNNNCAAGNRNN
|
0.036305931485634
|
4679
|
144
|
50
|
0.347222222222222
|
0.0106860440265014
|
|
TF
|
TF:M07379
|
Factor: c-Ets-2; motif: NNCTTCCTNNN
|
0.0365312971289607
|
3487
|
75
|
24
|
0.32
|
0.00688270719816461
|
|
TF
|
TF:M04849_1
|
Factor: RelA-p65; motif: AAASTCCC; match class: 1
|
0.0369478436105752
|
1170
|
31
|
7
|
0.225806451612903
|
0.00598290598290598
|
|
TF
|
TF:M12188_1
|
Factor: KLF17; motif: NGMCMCRCCCTN; match class: 1
|
0.0370467787644913
|
588
|
112
|
10
|
0.0892857142857143
|
0.0170068027210884
|
|
TF
|
TF:M08471
|
Factor: Fli-1:HOXB13; motif: NNCGGAARYNRTWAA
|
0.0375581637049886
|
5224
|
9
|
7
|
0.777777777777778
|
0.00133996937212864
|
|
TF
|
TF:M08881
|
Factor: ETSLIKE; motif: NACTTCCTNN
|
0.0376581925904836
|
3140
|
56
|
18
|
0.321428571428571
|
0.00573248407643312
|
|
TF
|
TF:M11209_1
|
Factor: AP-4; motif: ANCATATGNT; match class: 1
|
0.037847518381139
|
2803
|
72
|
20
|
0.277777777777778
|
0.00713521227256511
|
|
TF
|
TF:M08215
|
Factor: Elk-1:EVX1; motif: NNCGGWAATNNYMATTA
|
0.037847518381139
|
6557
|
69
|
35
|
0.507246376811594
|
0.00533780692389812
|
|
TF
|
TF:M00513
|
Factor: ATF3; motif: CBCTGACGTCANCS
|
0.0381514444127013
|
6386
|
56
|
29
|
0.517857142857143
|
0.00454118383964923
|
|
TF
|
TF:M01023
|
Factor: HSF1; motif: NTTCTRGAAVNTTCTYM
|
0.0381514444127013
|
2721
|
144
|
33
|
0.229166666666667
|
0.0121278941565601
|
|
TF
|
TF:M04360_1
|
Factor: HOXD11; motif: NNYMATAAAA; match class: 1
|
0.0381514444127013
|
1048
|
54
|
9
|
0.166666666666667
|
0.00858778625954199
|
|
TF
|
TF:M04254
|
Factor: FOXL1; motif: NRTAAAYAAACAN
|
0.0381514444127013
|
606
|
128
|
11
|
0.0859375
|
0.0181518151815182
|
|
TF
|
TF:M10565
|
Factor: Msx-1; motif: NCAATTAN
|
0.0381514444127013
|
2399
|
101
|
23
|
0.227722772277228
|
0.00958732805335557
|
|
TF
|
TF:M04292
|
Factor: BSX; motif: NYNATTAN
|
0.0381514444127013
|
2399
|
101
|
23
|
0.227722772277228
|
0.00958732805335557
|
|
TF
|
TF:M07270
|
Factor: TEF-3; motif: GNTATTTTT
|
0.0382809788453874
|
10215
|
79
|
54
|
0.683544303797468
|
0.0052863436123348
|
|
TF
|
TF:M10092
|
Factor: TEF-1; motif: RCATTCCWGNNN
|
0.0382809788453874
|
3195
|
27
|
11
|
0.407407407407407
|
0.00344287949921753
|
|
TF
|
TF:M00672
|
Factor: TEF; motif: ATGTTWAYATAA
|
0.0382809788453874
|
5921
|
73
|
34
|
0.465753424657534
|
0.00574227326465124
|
|
TF
|
TF:M05445
|
Factor: GATA-5; motif: TTAMCAR
|
0.0382809788453874
|
3283
|
54
|
18
|
0.333333333333333
|
0.00548279013097776
|
|
TF
|
TF:M03865_1
|
Factor: Blimp-1; motif: NRGRAAGKGAAAGK; match class: 1
|
0.0382809788453874
|
45
|
111
|
3
|
0.027027027027027
|
0.0666666666666667
|
|
TF
|
TF:M11360
|
Factor: c-MAF; motif: NTGCTGACNNNGCR
|
0.0382809788453874
|
221
|
117
|
6
|
0.0512820512820513
|
0.0271493212669683
|
|
TF
|
TF:M10081
|
Factor: STAT3; motif: NNYTTCCMRGAA
|
0.0385184121484572
|
3621
|
123
|
36
|
0.292682926829268
|
0.00994200497100249
|
|
TF
|
TF:M11445_1
|
Factor: Elf-1; motif: NANCAGGAAGTR; match class: 1
|
0.039203702334386
|
83
|
129
|
4
|
0.0310077519379845
|
0.0481927710843374
|
|
TF
|
TF:M12333
|
Factor: ZBTB20; motif: NNAAYGTATRKN
|
0.039203702334386
|
2707
|
123
|
29
|
0.235772357723577
|
0.0107129663834503
|
|
TF
|
TF:M08822_1
|
Factor: Msx-1; motif: WNGNAATTANV; match class: 1
|
0.0393654912626754
|
2272
|
6
|
4
|
0.666666666666667
|
0.00176056338028169
|
|
TF
|
TF:M11568
|
Factor: HNF-3alpha; motif: NNYTAWGTAAACAAAN
|
0.0395860765987651
|
2277
|
6
|
4
|
0.666666666666667
|
0.00175669740887132
|
|
TF
|
TF:M09625_1
|
Factor: GR; motif: RGNACANKNTGTNCY; match class: 1
|
0.0396590932011891
|
4205
|
57
|
22
|
0.385964912280702
|
0.00523186682520809
|
|
TF
|
TF:M09874
|
Factor: BCL-6; motif: NNCTTTCYAGGAA
|
0.0396590932011891
|
2807
|
119
|
29
|
0.243697478991597
|
0.0103313145707161
|
|
TF
|
TF:M04822
|
Factor: c-ets-1; motif: GGAGTTG
|
0.0397834945732107
|
5496
|
32
|
17
|
0.53125
|
0.00309315866084425
|
|
TF
|
TF:M10019
|
Factor: OLIG2; motif: CCAKCTGYTYNYNNNNNN
|
0.0397834945732107
|
3868
|
116
|
36
|
0.310344827586207
|
0.0093071354705274
|
|
TF
|
TF:M04750_1
|
Factor: GR; motif: RGRACATTNTGTYC; match class: 1
|
0.0402026688278395
|
1704
|
12
|
5
|
0.416666666666667
|
0.00293427230046948
|
|
TF
|
TF:M01653
|
Factor: HMGIY; motif: NGWWATTN
|
0.0402026688278395
|
6100
|
95
|
43
|
0.452631578947368
|
0.00704918032786885
|
|
TF
|
TF:M03886
|
Factor: Sox-2; motif: NNCCWTTGTTNTKN
|
0.0402026688278395
|
993
|
6
|
3
|
0.5
|
0.00302114803625378
|
|
TF
|
TF:M07301
|
Factor: NFATc4; motif: NGGAAAAN
|
0.0404646854511435
|
4070
|
66
|
24
|
0.363636363636364
|
0.0058968058968059
|
|
TF
|
TF:M08461
|
Factor: Erm:EVX1; motif: NNCGGAAATKR
|
0.0406712497456048
|
3396
|
19
|
9
|
0.473684210526316
|
0.00265017667844523
|
|
TF
|
TF:M03545
|
Factor: c-Rel; motif: NGGGAATYTCCN
|
0.0407737983454631
|
5530
|
53
|
25
|
0.471698113207547
|
0.00452079566003617
|
|
TF
|
TF:M10042_1
|
Factor: PR; motif: RGNACATTYTGTNCTN; match class: 1
|
0.0407737983454631
|
998
|
12
|
4
|
0.333333333333333
|
0.00400801603206413
|
|
TF
|
TF:M01012
|
Factor: HNF3; motif: NNNNNTRTTTRYTYWNKN
|
0.0414623599452486
|
2298
|
22
|
8
|
0.363636363636364
|
0.00348128807658834
|
|
TF
|
TF:M01884_1
|
Factor: IRF-7; motif: AAGWGAA; match class: 1
|
0.0414623599452486
|
4556
|
135
|
46
|
0.340740740740741
|
0.0100965759438104
|
|
TF
|
TF:M10640_1
|
Factor: CSX; motif: NCCACTTRAN; match class: 1
|
0.0414623599452486
|
1237
|
10
|
4
|
0.4
|
0.00323362974939369
|
|
TF
|
TF:M08888
|
Factor: MAFB; motif: NTCAGCN
|
0.0415688880215711
|
10140
|
140
|
89
|
0.635714285714286
|
0.00877712031558185
|
|
TF
|
TF:M08907_1
|
Factor: AR; motif: AGAACANNNTGTTCT; match class: 1
|
0.0418810844549913
|
2176
|
50
|
13
|
0.26
|
0.00597426470588235
|
|
TF
|
TF:M01808
|
Factor: NMYC; motif: CAYCTG
|
0.0418810844549913
|
12944
|
59
|
49
|
0.830508474576271
|
0.00378553770086527
|
|
TF
|
TF:M03813_1
|
Factor: BRN1; motif: HAATGCN; match class: 1
|
0.0418871834086463
|
7077
|
72
|
38
|
0.527777777777778
|
0.00536950685318638
|
|
TF
|
TF:M04802
|
Factor: c-Fos; motif: ACTCACCA
|
0.0421081593743279
|
7386
|
128
|
64
|
0.5
|
0.00866504197129705
|
|
TF
|
TF:M03872
|
Factor: HNF-1beta; motif: GTTAATYATTAACY
|
0.0421306019148706
|
4145
|
76
|
27
|
0.355263157894737
|
0.00651387213510253
|
|
TF
|
TF:M03579
|
Factor: PEA3; motif: AGGAAGT
|
0.042164352819053
|
6837
|
58
|
31
|
0.53448275862069
|
0.00453415240602603
|
|
TF
|
TF:M06481
|
Factor: ZNF433; motif: AAMCTGATGMCA
|
0.042164352819053
|
44
|
121
|
3
|
0.0247933884297521
|
0.0681818181818182
|
|
TF
|
TF:M07064
|
Factor: STAT1; motif: TTCNRGGAAN
|
0.042164352819053
|
2606
|
123
|
28
|
0.227642276422764
|
0.0107444359171144
|
|
TF
|
TF:M07382
|
Factor: Fli-1; motif: MGGAAGT
|
0.042164352819053
|
6837
|
58
|
31
|
0.53448275862069
|
0.00453415240602603
|
|
TF
|
TF:M08482
|
Factor: FOXO1A:HOXA10; motif: RWMAACANCRTWAA
|
0.042164352819053
|
4206
|
54
|
21
|
0.388888888888889
|
0.00499286733238231
|
|
TF
|
TF:M01870
|
Factor: C-ets-1; motif: AGGAAGN
|
0.042164352819053
|
6837
|
58
|
31
|
0.53448275862069
|
0.00453415240602603
|
|
TF
|
TF:M02278
|
Factor: SPI1; motif: AGGAAGT
|
0.042164352819053
|
6837
|
58
|
31
|
0.53448275862069
|
0.00453415240602603
|
|
TF
|
TF:M01595
|
Factor: STAT3; motif: NNTTCCRGGAANNNNN
|
0.042164352819053
|
7736
|
88
|
48
|
0.545454545454545
|
0.0062047569803516
|
|
TF
|
TF:M11549
|
Factor: HFH-1; motif: YTANRTAAACWN
|
0.0421675672853781
|
839
|
96
|
11
|
0.114583333333333
|
0.0131108462455304
|
|
TF
|
TF:M07421
|
Factor: HOXC-8; motif: GAATWAYARN
|
0.0427998295420725
|
1504
|
55
|
11
|
0.2
|
0.00731382978723404
|
|
TF
|
TF:M02116_1
|
Factor: Sox-10; motif: NACAAWG; match class: 1
|
0.042821831044917
|
8039
|
104
|
57
|
0.548076923076923
|
0.00709043413359871
|
|
TF
|
TF:M00340_1
|
Factor: c-Ets; motif: KRCAGGAARTRNKT; match class: 1
|
0.042821831044917
|
906
|
116
|
13
|
0.112068965517241
|
0.0143487858719647
|
|
TF
|
TF:M00432
|
Factor: TTF1; motif: ASTCAAGTRK
|
0.0433337378105857
|
5394
|
122
|
48
|
0.39344262295082
|
0.00889877641824249
|
|
TF
|
TF:M11383_1
|
Factor: ESE-1; motif: ACCAGGAAGTANNNNNNNAAWAA; match class: 1
|
0.043338394495662
|
589
|
80
|
8
|
0.1
|
0.0135823429541596
|
|
TF
|
TF:M10074
|
Factor: Spi-B; motif: NRAAAGAGGAAGTGARA
|
0.043338394495662
|
1611
|
142
|
22
|
0.154929577464789
|
0.0136561142147734
|
|
TF
|
TF:M00926
|
Factor: AP-1; motif: TGAGTCAN
|
0.0437029413379206
|
924
|
52
|
8
|
0.153846153846154
|
0.00865800865800866
|
|
TF
|
TF:M01267
|
Factor: Fra-1; motif: TGAGTCAN
|
0.0437029413379206
|
924
|
52
|
8
|
0.153846153846154
|
0.00865800865800866
|
|
TF
|
TF:M00423
|
Factor: FOXJ2; motif: AYMATAATATTTKN
|
0.0437029413379206
|
6970
|
55
|
30
|
0.545454545454545
|
0.00430416068866571
|
|
TF
|
TF:M01307
|
Factor: Oct3; motif: NATGCAANNN
|
0.0437029413379206
|
3986
|
23
|
11
|
0.478260869565217
|
0.00275965880582037
|
|
TF
|
TF:M11290_1
|
Factor: ATF-3; motif: NATGATGTCATN; match class: 1
|
0.0437029413379206
|
1872
|
79
|
16
|
0.20253164556962
|
0.00854700854700855
|
|
TF
|
TF:M08926
|
Factor: JUNB; motif: ATGACTCA
|
0.0437029413379206
|
924
|
52
|
8
|
0.153846153846154
|
0.00865800865800866
|
|
TF
|
TF:M10713_1
|
Factor: HOXD4; motif: RTCRTTAN; match class: 1
|
0.0437029413379206
|
616
|
77
|
8
|
0.103896103896104
|
0.012987012987013
|
|
TF
|
TF:M04305
|
Factor: DMBX1; motif: NNGGATTANN
|
0.0437029413379206
|
5134
|
60
|
26
|
0.433333333333333
|
0.00506427736657577
|
|
TF
|
TF:M04054_1
|
Factor: NF-kappaB; motif: AGGGGAWTCCCCT; match class: 1
|
0.0439026259808883
|
1295
|
30
|
7
|
0.233333333333333
|
0.00540540540540541
|
|
TF
|
TF:M09635
|
Factor: MafK; motif: NAAWNTGCTGASTCAGCANWWN
|
0.0439026259808883
|
68
|
81
|
3
|
0.037037037037037
|
0.0441176470588235
|
|
TF
|
TF:M04415
|
Factor: NKX6-1; motif: NTMATTAA
|
0.04391264849858
|
1846
|
66
|
14
|
0.212121212121212
|
0.0075839653304442
|
|
TF
|
TF:M00095_1
|
Factor: CDP; motif: CCAATAATCGAT; match class: 1
|
0.0442218110715789
|
2983
|
99
|
26
|
0.262626262626263
|
0.00871605766007375
|
|
TF
|
TF:M00205
|
Factor: GR; motif: GGTACAANNTGTYCTK
|
0.0443972012059103
|
6653
|
55
|
29
|
0.527272727272727
|
0.00435893581842778
|
|
TF
|
TF:M11908_1
|
Factor: POU3F2; motif: TAATKAGNNNNTAATKA; match class: 1
|
0.0444982264234007
|
1278
|
23
|
6
|
0.260869565217391
|
0.00469483568075117
|
|
TF
|
TF:M11696_1
|
Factor: MEF-2B1; motif: CCNWATWWRG; match class: 1
|
0.0447789378692664
|
898
|
7
|
3
|
0.428571428571429
|
0.00334075723830735
|
|
TF
|
TF:M11983
|
Factor: NFATc2; motif: NTTTCCGCGGAAAN
|
0.0454119805208158
|
8048
|
69
|
40
|
0.579710144927536
|
0.00497017892644135
|
|
TF
|
TF:M07065
|
Factor: STAT3; motif: TTCYGGGAAN
|
0.0459454853858555
|
4948
|
123
|
45
|
0.365853658536585
|
0.00909458367016977
|
|
TF
|
TF:M07104
|
Factor: JunD; motif: NRTGASTCATN
|
0.0459454853858555
|
2675
|
52
|
15
|
0.288461538461538
|
0.00560747663551402
|
|
TF
|
TF:M00224_1
|
Factor: STAT1; motif: NNNSANTTCCGGGAANTGNSN; match class: 1
|
0.0459454853858555
|
2710
|
98
|
24
|
0.244897959183673
|
0.00885608856088561
|
|
TF
|
TF:M10814
|
Factor: HOXC11; motif: NGYMATAAAAN
|
0.0459454853858555
|
756
|
64
|
8
|
0.125
|
0.0105820105820106
|
|
TF
|
TF:M01884
|
Factor: IRF-7; motif: AAGWGAA
|
0.0459454853858555
|
10868
|
121
|
82
|
0.677685950413223
|
0.00754508649245491
|
|
TF
|
TF:M04224
|
Factor: NRL; motif: NNNNTGCTGAC
|
0.046013720666635
|
4667
|
123
|
43
|
0.349593495934959
|
0.00921362759802871
|
|
TF
|
TF:M06005
|
Factor: ZSCAN11; motif: NYMKAGGAAASA
|
0.046013720666635
|
311
|
90
|
6
|
0.0666666666666667
|
0.0192926045016077
|
|
TF
|
TF:M03882_1
|
Factor: RelB:p50; motif: RGAAANTCCCYNNHGC; match class: 1
|
0.046013720666635
|
714
|
115
|
11
|
0.0956521739130435
|
0.015406162464986
|
|
TF
|
TF:M04233
|
Factor: FOXC1; motif: NGTMAATATTKACN
|
0.046013720666635
|
114
|
103
|
4
|
0.0388349514563107
|
0.0350877192982456
|
|
TF
|
TF:M09839
|
Factor: ATF4; motif: ATTGCATCAT
|
0.046013720666635
|
2145
|
46
|
12
|
0.260869565217391
|
0.00559440559440559
|
|
TF
|
TF:M07417
|
Factor: Elf-1; motif: NNCAGGAAGNNN
|
0.046013720666635
|
1774
|
107
|
19
|
0.177570093457944
|
0.0107102593010147
|
|
TF
|
TF:M02377_1
|
Factor: ESR2; motif: NNRGGTCANNNTGMCCTN; match class: 1
|
0.046063088387841
|
690
|
9
|
3
|
0.333333333333333
|
0.00434782608695652
|
|
TF
|
TF:M11334
|
Factor: ATF-4; motif: GGATGATGTCATCC
|
0.0466338201104689
|
6386
|
16
|
11
|
0.6875
|
0.00172251800814281
|
|
TF
|
TF:M08890
|
Factor: MYB; motif: NNAACTGN
|
0.0466338201104689
|
1355
|
139
|
19
|
0.136690647482014
|
0.0140221402214022
|
|
TF
|
TF:M07268_1
|
Factor: Sox-4; motif: BCWTTGT; match class: 1
|
0.0466540395351024
|
1086
|
66
|
10
|
0.151515151515152
|
0.00920810313075507
|
|
TF
|
TF:M11431
|
Factor: PEA3; motif: NTCGTAAATGCA
|
0.0468521669398452
|
10559
|
86
|
59
|
0.686046511627907
|
0.00558765034567667
|
|
TF
|
TF:M01308_1
|
Factor: Sox-4; motif: AACAAWGR; match class: 1
|
0.0469078232840993
|
107
|
54
|
3
|
0.0555555555555556
|
0.0280373831775701
|
|
TF
|
TF:M02080
|
Factor: Isl2; motif: YTAAGTG
|
0.0470019115088098
|
6041
|
144
|
60
|
0.416666666666667
|
0.0099321304419798
|
|
TF
|
TF:M09959_1
|
Factor: IRF-4; motif: NAANGRGGAASTGAAASN; match class: 1
|
0.0470960254768766
|
395
|
72
|
6
|
0.0833333333333333
|
0.0151898734177215
|
|
TF
|
TF:M09618
|
Factor: HNF-3alpha; motif: NTRTTTACWYWNN
|
0.0474364637464237
|
2353
|
59
|
15
|
0.254237288135593
|
0.00637484062898428
|
|
TF
|
TF:M04078
|
Factor: POU3F2; motif: NWTGCATAAWTTA
|
0.0474364637464237
|
3944
|
62
|
22
|
0.354838709677419
|
0.00557809330628803
|
|
TF
|
TF:M04476_1
|
Factor: NR3C1; motif: NRGWACAYNRTGTWCYN; match class: 1
|
0.0476264301129285
|
5349
|
121
|
47
|
0.388429752066116
|
0.0087866891007665
|
|
TF
|
TF:M04399
|
Factor: MEOX2; motif: NSTAATTANN
|
0.0478085977629644
|
3837
|
103
|
32
|
0.310679611650485
|
0.00833984884023977
|
|
TF
|
TF:M00158
|
Factor: COUP-TF,; motif: TGAMCTTTGMMCYT
|
0.0480679096736997
|
2215
|
133
|
26
|
0.195488721804511
|
0.0117381489841986
|
|
TF
|
TF:M00955
|
Factor: GR; motif: NNNNNNNGKACNNNNTGTTCTNNNNNN
|
0.0480679096736997
|
4958
|
36
|
17
|
0.472222222222222
|
0.00342880193626462
|
|
TF
|
TF:M08907
|
Factor: AR; motif: AGAACANNNTGTTCT
|
0.0480679096736997
|
2256
|
50
|
13
|
0.26
|
0.00576241134751773
|
|
TF
|
TF:M08369
|
Factor: HOXD12:PEA3; motif: RCCGGAAGTAATAAAM
|
0.0480679096736997
|
4596
|
6
|
5
|
0.833333333333333
|
0.00108790252393386
|
|
TF
|
TF:M03996
|
Factor: SPIC; motif: NAAAAGMGGAAGTA
|
0.0480679096736997
|
670
|
124
|
11
|
0.0887096774193548
|
0.0164179104477612
|
|
TF
|
TF:M01292
|
Factor: HOXA13; motif: ATAAMA
|
0.0480679096736997
|
13416
|
79
|
65
|
0.822784810126582
|
0.00484496124031008
|
|
TF
|
TF:M04340
|
Factor: HOXA10; motif: NGYMATAAAAN
|
0.0480679096736997
|
1978
|
21
|
7
|
0.333333333333333
|
0.00353892821031345
|
|
TF
|
TF:M04077
|
Factor: POU3F1; motif: WTGMATAAWTNA
|
0.0480679096736997
|
2836
|
105
|
26
|
0.247619047619048
|
0.00916784203102962
|
|
TF
|
TF:M08931
|
Factor: JUND:FRA-2; motif: NATGACTCAT
|
0.0480679096736997
|
1410
|
52
|
10
|
0.192307692307692
|
0.00709219858156028
|
|
TF
|
TF:M10541
|
Factor: SUHW1; motif: TCTCTCCAGTRTGAATTCTCTGAT
|
0.0480679096736997
|
7303
|
80
|
42
|
0.525
|
0.00575106120772285
|
|
TF
|
TF:M08976_1
|
Factor: SRY; motif: AACAATNR; match class: 1
|
0.0482706269799117
|
8093
|
63
|
37
|
0.587301587301587
|
0.00457185221796614
|
|
TF
|
TF:M00746
|
Factor: Elf-1; motif: RNWMBAGGAART
|
0.0482706269799117
|
3365
|
72
|
22
|
0.305555555555556
|
0.00653789004457652
|
|
TF
|
TF:M02025_1
|
Factor: MEF-2C; motif: TATTTWT; match class: 1
|
0.0482762056652641
|
7743
|
25
|
17
|
0.68
|
0.00219553144775927
|
|
TF
|
TF:M07047_1
|
Factor: Lhx2; motif: WATTAN; match class: 1
|
0.0482762056652641
|
4624
|
6
|
5
|
0.833333333333333
|
0.00108131487889273
|
|
TF
|
TF:M08003_1
|
Factor: POU6F1; motif: ANTAATTANT; match class: 1
|
0.0484830983536778
|
525
|
12
|
3
|
0.25
|
0.00571428571428571
|
|
TF
|
TF:M08003
|
Factor: POU6F1; motif: ANTAATTANT
|
0.0484830983536778
|
525
|
12
|
3
|
0.25
|
0.00571428571428571
|
|
TF
|
TF:M00925
|
Factor: AP-1; motif: NTGACTCAN
|
0.0485683584394179
|
2554
|
134
|
29
|
0.216417910447761
|
0.0113547376664056
|
|
TF
|
TF:M11983_1
|
Factor: NFATc2; motif: NTTTCCGCGGAAAN; match class: 1
|
0.0489270649617097
|
7294
|
69
|
37
|
0.536231884057971
|
0.00507266246229778
|
|
TF
|
TF:M07204_1
|
Factor: AR; motif: ARGAACANNNTGTNC; match class: 1
|
0.0489270649617097
|
963
|
52
|
8
|
0.153846153846154
|
0.0083073727933541
|
|
TF
|
TF:M01869
|
Factor: C/EBPgamma; motif: NRTKRSRMAAKN
|
0.0491081991016935
|
3194
|
99
|
27
|
0.272727272727273
|
0.0084533500313087
|
|
TF
|
TF:M04453_1
|
Factor: AR; motif: RGGWACAYNGTGTWCYN; match class: 1
|
0.0491413086131068
|
3794
|
57
|
20
|
0.350877192982456
|
0.00527148128624143
|
|
TF
|
TF:M02108
|
Factor: CSX; motif: NNCACTTGNRN
|
0.0492143858748774
|
7061
|
135
|
64
|
0.474074074074074
|
0.00906387197280838
|
|
TF
|
TF:M08575
|
Factor: GCMa:C/EBPbeta; motif: ATGCGGGTNNRTTGCGCAAY
|
0.0492143858748774
|
7026
|
17
|
12
|
0.705882352941177
|
0.00170794192997438
|
|
TF
|
TF:M09725_1
|
Factor: DREF; motif: CTYYCWCTTCCY; match class: 1
|
0.0492508454625437
|
2061
|
136
|
25
|
0.183823529411765
|
0.0121300339640951
|
|
TF
|
TF:M10673
|
Factor: Emx-2; motif: YTAATTAN
|
0.0493452098728638
|
3224
|
103
|
28
|
0.271844660194175
|
0.0086848635235732
|
|
TF
|
TF:M04446
|
Factor: VAX1; motif: YTAATTAN
|
0.0493452098728638
|
3224
|
103
|
28
|
0.271844660194175
|
0.0086848635235732
|
|
TF
|
TF:M10021
|
Factor: Otx2; motif: NRRGGATTARN
|
0.0493452098728638
|
1679
|
59
|
12
|
0.203389830508475
|
0.00714711137581894
|
|
TF
|
TF:M11909_1
|
Factor: POU3F2; motif: NTATGCWAATKAG; match class: 1
|
0.0493452098728638
|
1184
|
72
|
11
|
0.152777777777778
|
0.00929054054054054
|
|
TF
|
TF:M01149
|
Factor: DMRT4; motif: AATGTADCAAWTT
|
0.0494545639871605
|
1842
|
61
|
13
|
0.213114754098361
|
0.00705754614549403
|
|
TF
|
TF:M00776_1
|
Factor: SREBP; motif: VNNVTCACCCYA; match class: 1
|
0.0494696119461393
|
1289
|
76
|
12
|
0.157894736842105
|
0.00930954228083786
|
|
TF
|
TF:M11632_1
|
Factor: Sox-10; motif: AACAATRGNCYATTGTT; match class: 1
|
0.0497356413315427
|
4639
|
67
|
26
|
0.388059701492537
|
0.00560465617589998
|
|
WP
|
WP:WP619
|
Type II interferon signaling (IFNG)
|
3.06220195272461e-16
|
37
|
68
|
13
|
0.191176470588235
|
0.351351351351351
|
|
WP
|
WP:WP4197
|
The human immune response to tuberculosis
|
4.89265162031772e-13
|
23
|
115
|
11
|
0.0956521739130435
|
0.478260869565217
|
|
WP
|
WP:WP4880
|
Host-pathogen interaction of human corona viruses - Interferon induction
|
8.37497563492182e-08
|
33
|
84
|
8
|
0.0952380952380952
|
0.242424242424242
|
|
WP
|
WP:WP4868
|
Type I Interferon Induction and Signaling During SARS-CoV-2 Infection
|
3.69960136800606e-06
|
29
|
63
|
6
|
0.0952380952380952
|
0.206896551724138
|
|
WP
|
WP:WP5039
|
SARS-CoV-2 Innate Immunity Evasion and Cell-specific immune response
|
1.95508104475663e-05
|
68
|
84
|
8
|
0.0952380952380952
|
0.117647058823529
|
|
WP
|
WP:WP4341
|
Non-genomic actions of 1,25 dihydroxyvitamin D3
|
0.000354835319487682
|
70
|
35
|
5
|
0.142857142857143
|
0.0714285714285714
|
|
WP
|
WP:WP4705
|
Pathways of nucleic acid metabolism and innate immune sensing
|
0.000461369817014912
|
16
|
84
|
4
|
0.0476190476190476
|
0.25
|
|
WP
|
WP:WP4912
|
SARS coronavirus and innate immunity
|
0.00491939753750522
|
31
|
84
|
4
|
0.0476190476190476
|
0.129032258064516
|
|
WP
|
WP:WP4877
|
Host-pathogen interaction of human corona viruses - MAPK signaling
|
0.0117886016179128
|
36
|
41
|
3
|
0.0731707317073171
|
0.0833333333333333
|
|
WP
|
WP:WP183
|
Proteasome Degradation
|
0.0206594219133776
|
64
|
104
|
5
|
0.0480769230769231
|
0.078125
|
|
WP
|
WP:WP3929
|
Chemokine signaling pathway
|
0.0332333609996534
|
165
|
91
|
7
|
0.0769230769230769
|
0.0424242424242424
|
|
WP
|
WP:WP2328
|
Allograft Rejection
|
0.049185345581961
|
88
|
104
|
5
|
0.0480769230769231
|
0.0568181818181818
|